Overview
Binder specifications define the protein binder properties (modality, structure, design regions) that will be used to generate candidates.Creating a New Binder Specification
- From your project dashboard, click ”+ Add” in the Binder Specifications section.
-
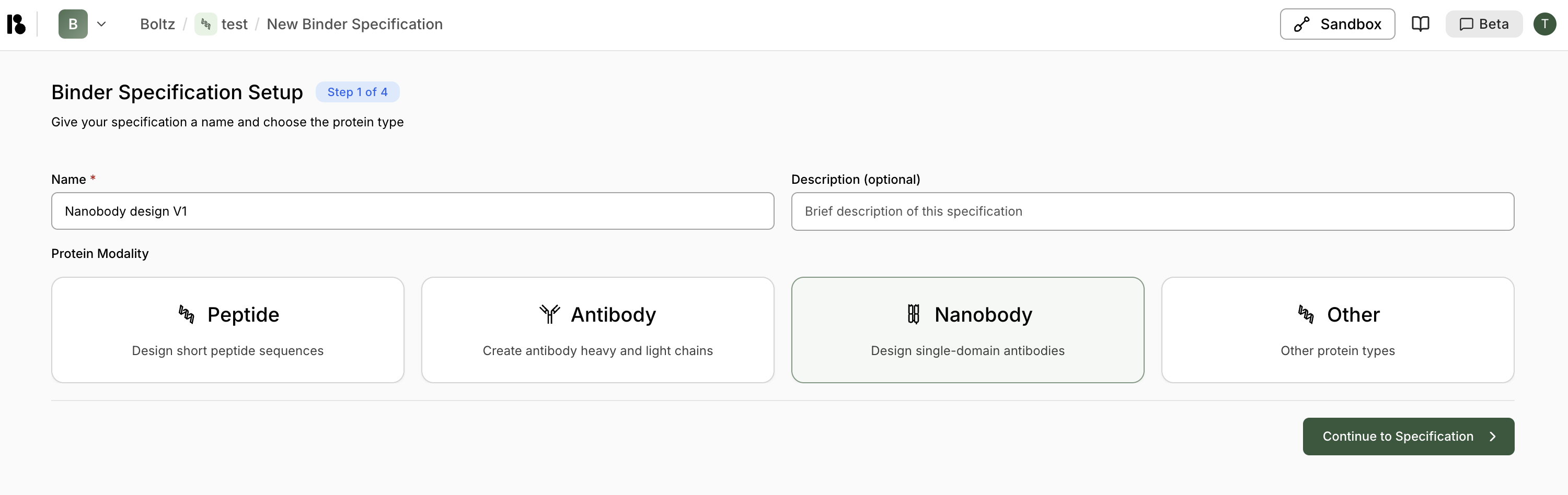
Step 1: Setup - Give your binder specification a name and choose the protein modality:
Peptide
Design short peptide sequencesAntibody
Create antibody heavy and light chainsNanobody
Design single-domain antibodiesOther
Other protein types - Click ”> Continue to Specification”.
Starting Specification
Option 1: Structure-Based Specification
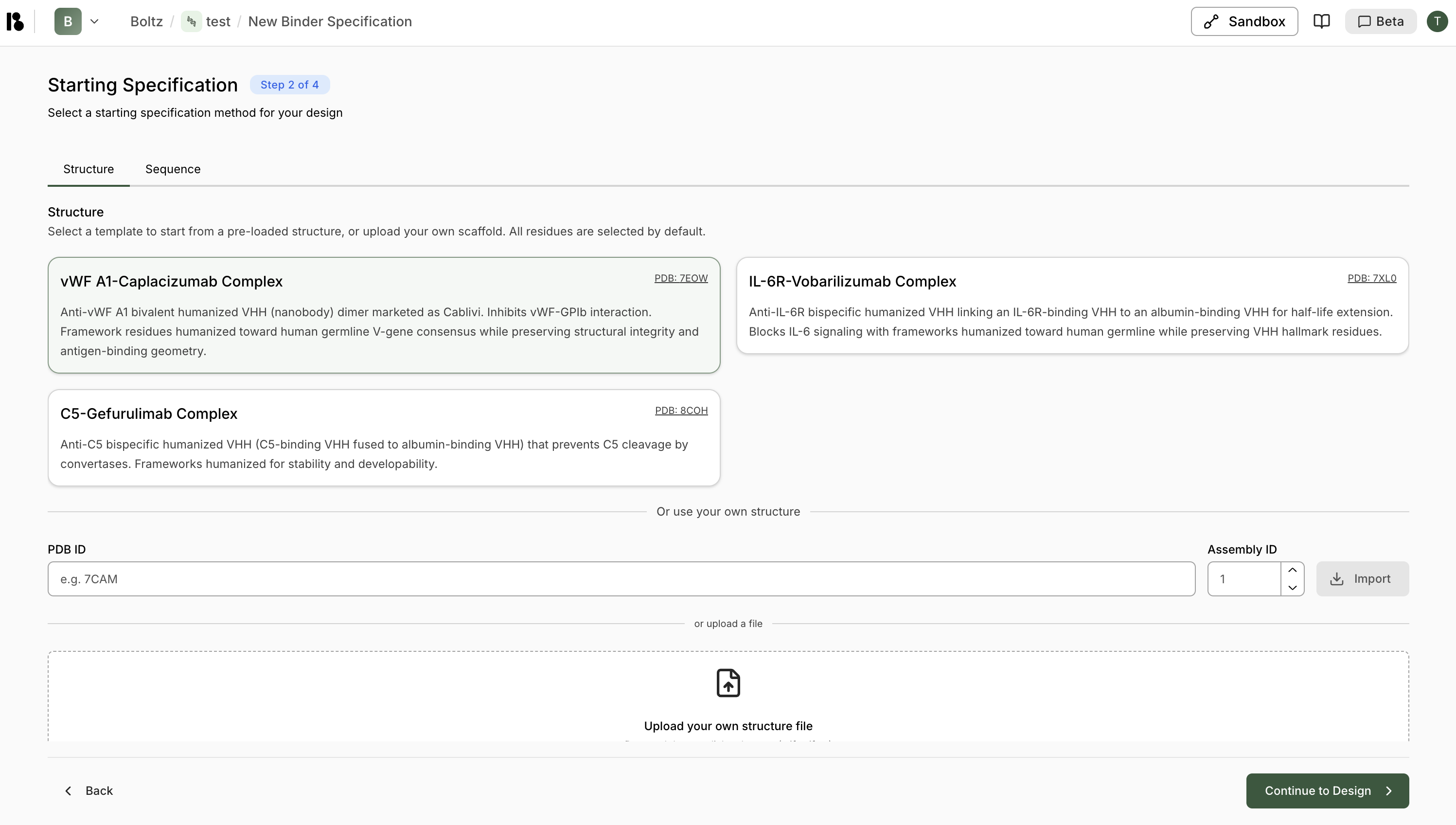
- Choose from pre-loaded structures (e.g., vWF A1-Caplacizumab Complex, IL-6R-Vobarilizumab Complex)
- Each template shows PDB ID and description
- Enter a PDB ID and Assembly ID (default: 1), then click “Import”
- Or upload a structure file (.cif, .cif.gz, etc.)
Option 2: Sequence-Based Specification
Click the “Sequence” tab to add sequences directly instead of using a structure.Select Residues
- Recommended: ≤300 residues
- Use CDR1, CDR2, CDR3 quick-select buttons for antibodies/nanobodies
- Or manually select by clicking and dragging on the sequence
- Selected residues are highlighted in green
- The 3D structure updates to show selected regions
- Chothia (default) or Kabat numbering for antibodies/nanobodies
- Switch between schemes using the tabs above the sequence
Design Configuration
Configure design motifs and constraints: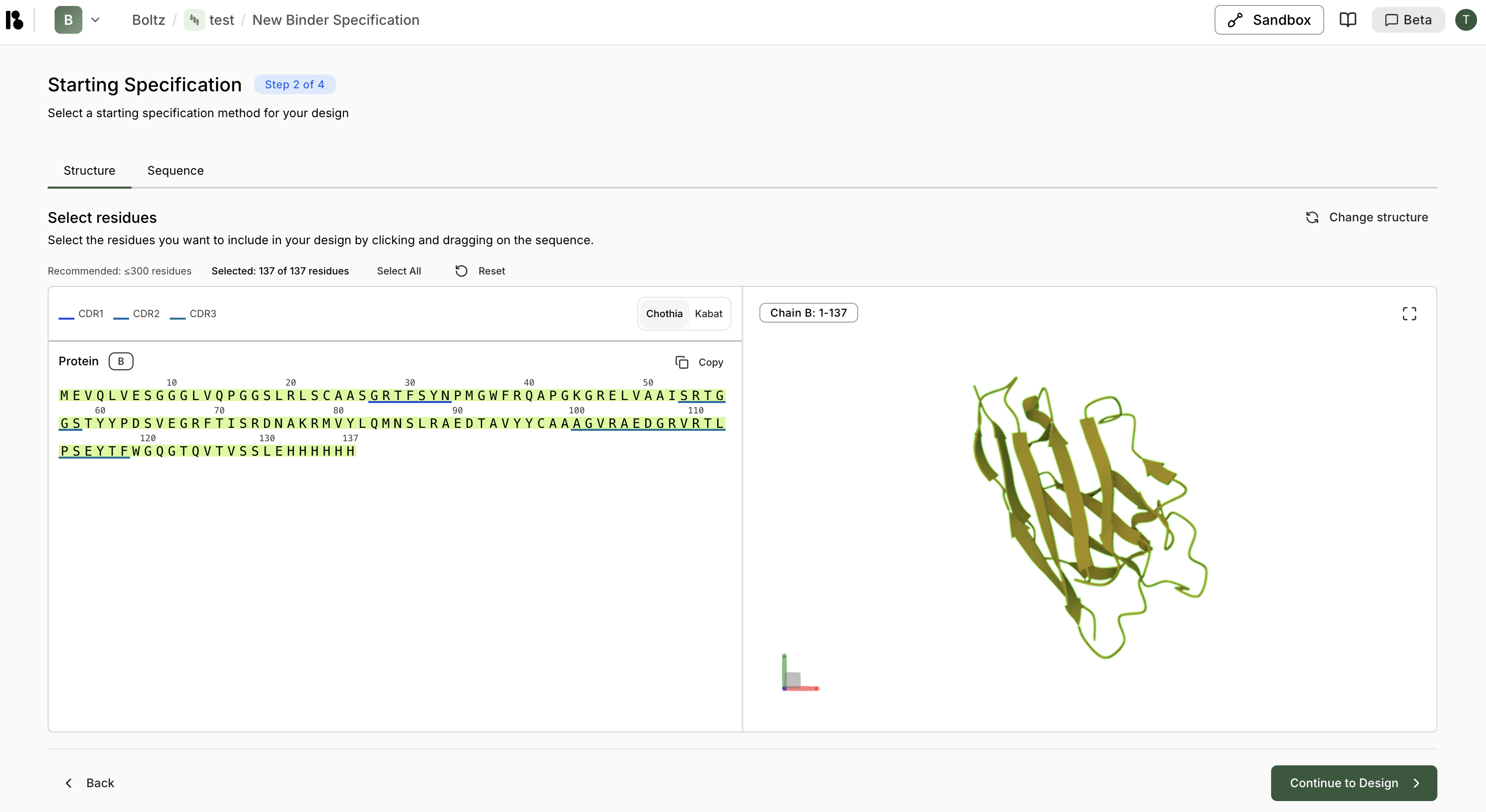
Excluded Amino Acids
Specify amino acids that will not be used in designed regions (e.g., exclude Cysteine to prevent unwanted disulfide bonds).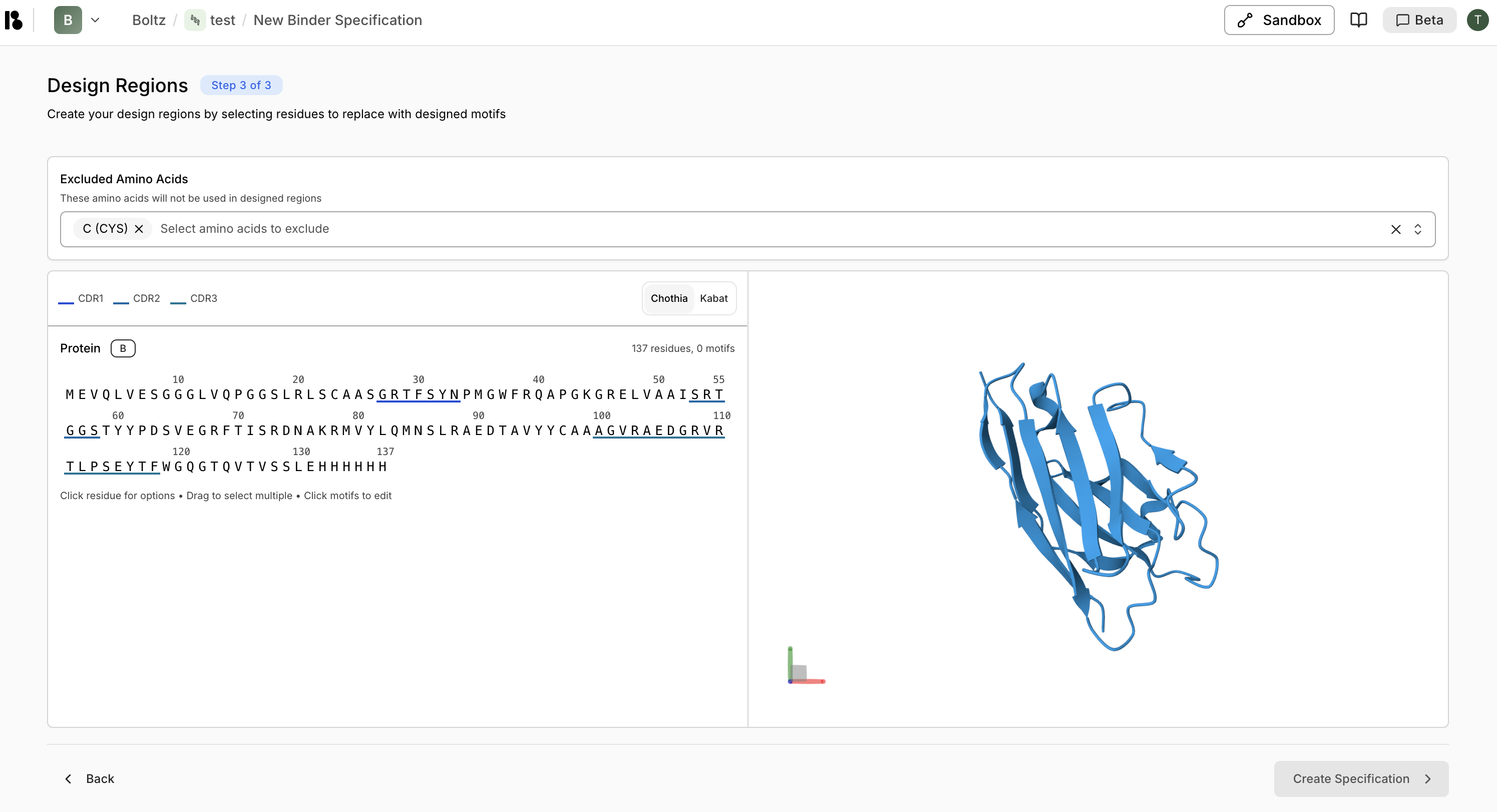
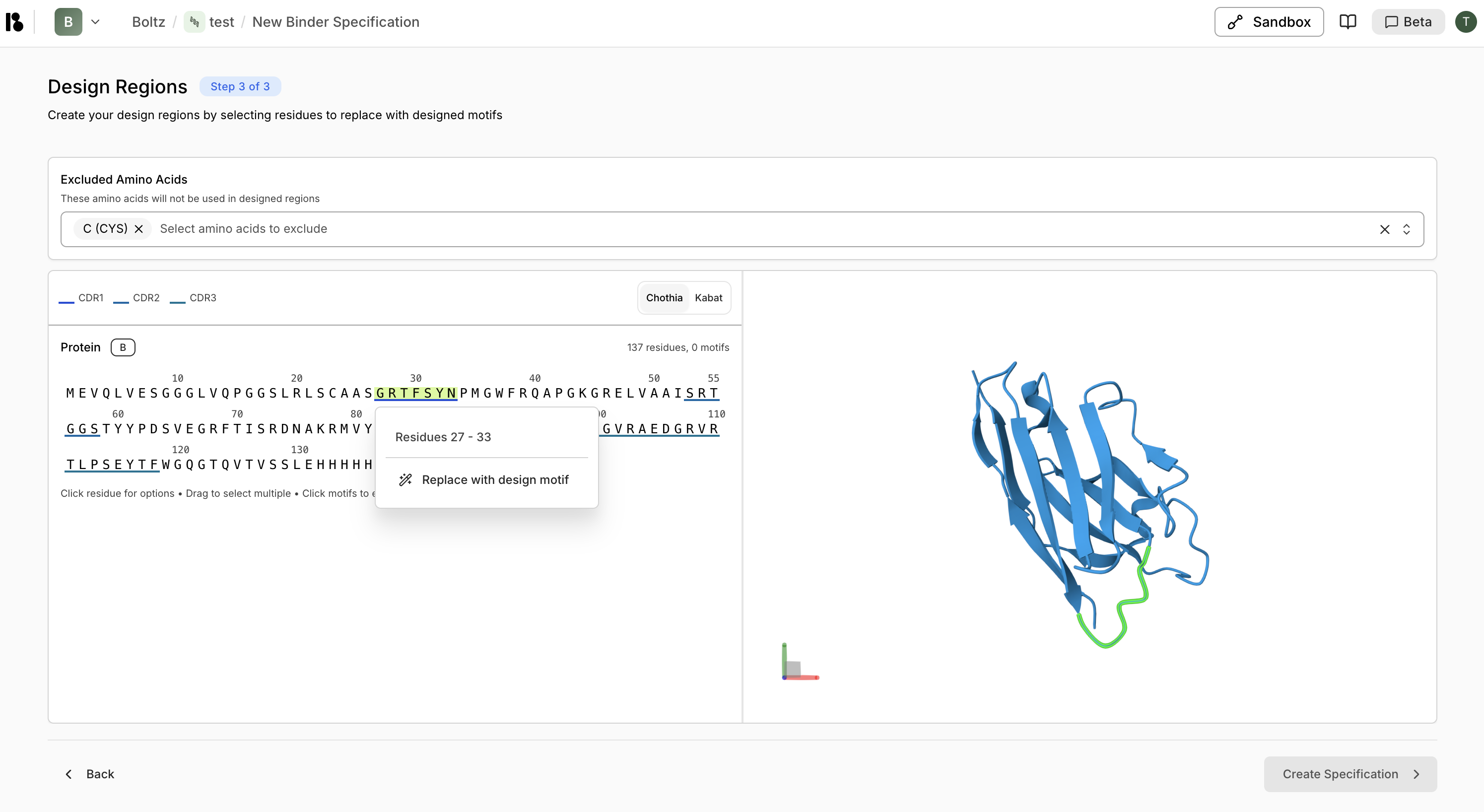
Create Design Motifs
- Select residues on the sequence or 3D structure
- Right-click or use the context menu to “Replace with design motif”
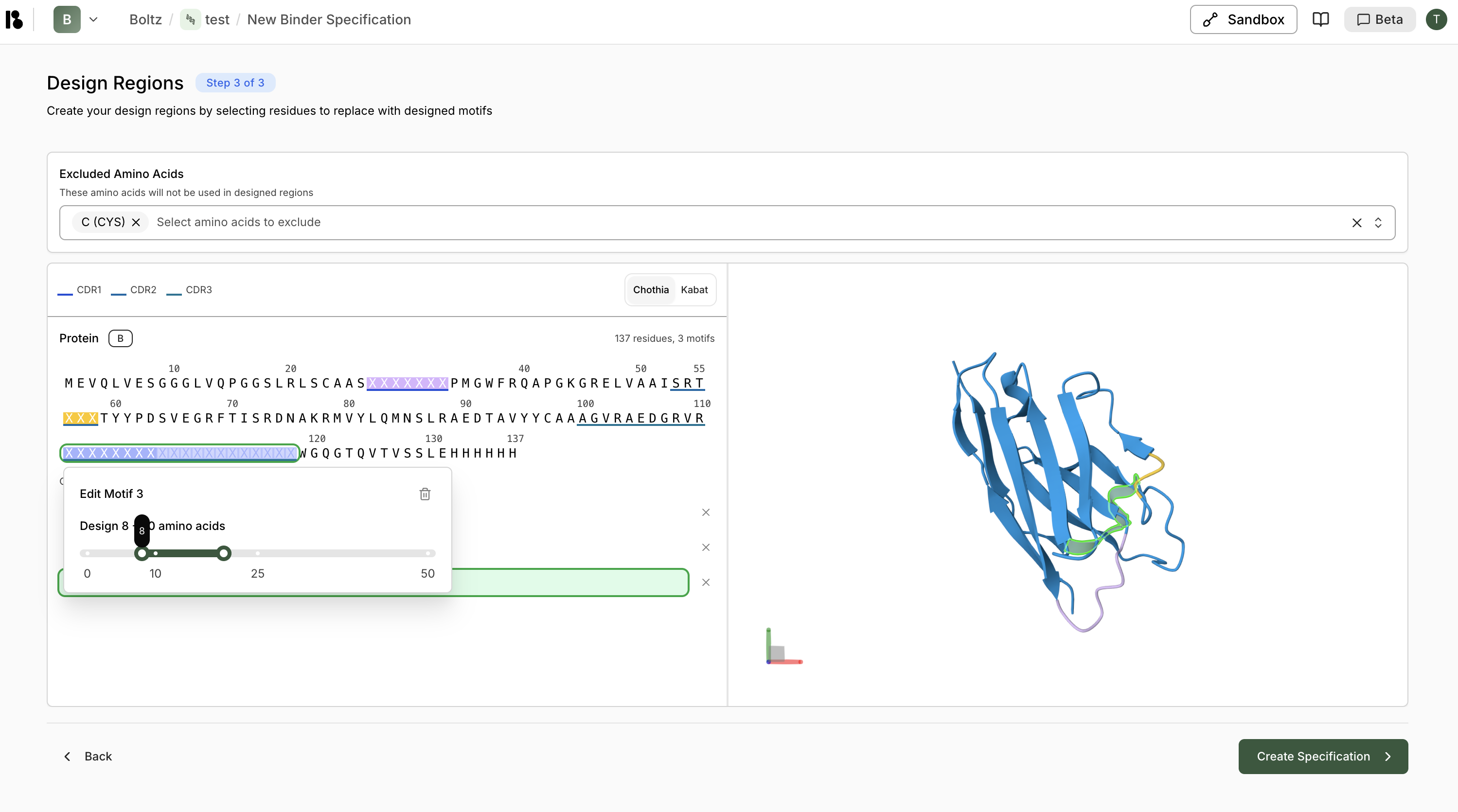
- For CDR regions, you can specify:
- Fixed length - Replace with same number of residues
- Variable length - Specify a range (e.g., 10-25 residues for CDR3)
Motif Management
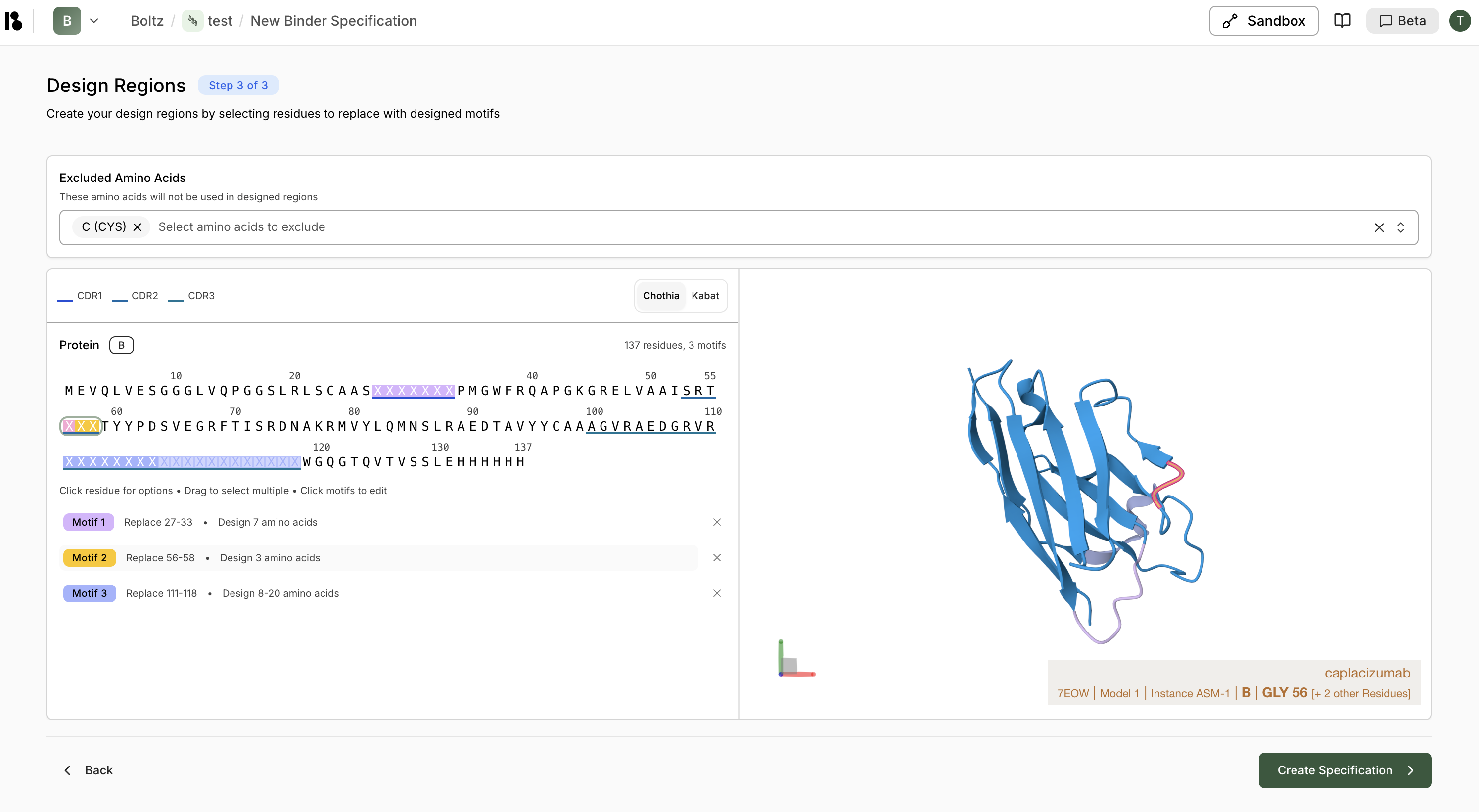
- Each motif is color-coded and shown in the sequence viewer
- Click a motif to edit it
- Click the X on a motif to remove it
- Summary shows total residues and number of motifs
After Binder Specification Creation
Once your binder specification is created, you’ll see options to:- Generate binders with AI - Start a virtual screen using this binder specification
- Create Another Target - Add another target to design binders against
- Skip for now - Continue later