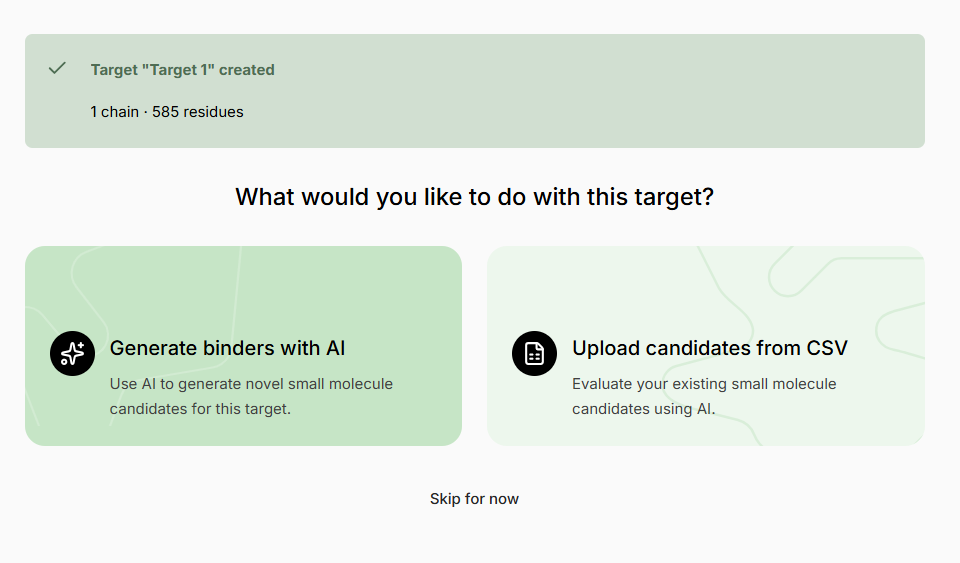
Creating an Experiment
Follow the instruction and select + Create New Experiment fromprompts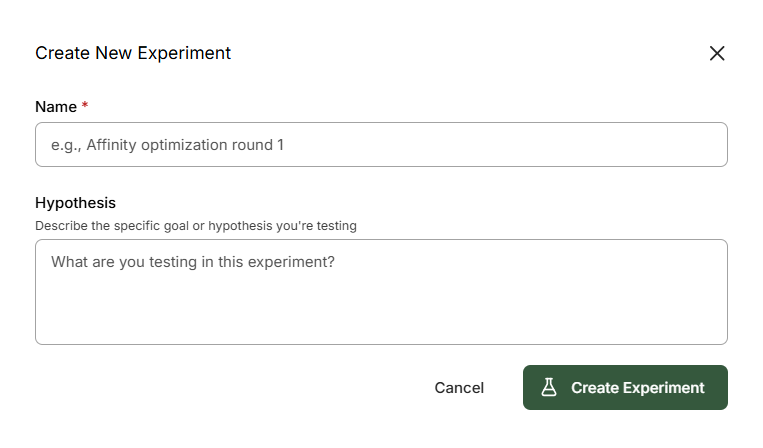
- ❌ Poor Example
- ✅ Good Example
Name: Experiment 1Hypothesis: Molecules for screening
Generative Virtual Screen
A Generative screen uses generative AI models and active learning to search molecular space and find binders for the selected Target, optimizing the predicted binding confidence..png?fit=max&auto=format&n=01Dj948NLM75wx38&q=85&s=0b0cb58ddcc8944cc9d508b57a333034)
Parameters
Target Selection
Target Selection
If multiple Targets are added to the Design Project, select the target you want to optimize affinity for.
Chemical Space
Chemical Space
By default, a generative screen searches within the expanded ~75B Enamine REAL Space. This ensures all output molecules are theoretically synthesizable. Alternatively, choose not to apply any chemical-space filter.
Molecule Structure Filtering
Molecule Structure Filtering
Normal Filtering (default): Curated filtering excluding most problematic molecules but allowing some motifs (phenols, anilines, aryl halides, esters) that can be optimized away in later design cycles.Extra Filtering: Extends to include functional groups with chemical stability issues.Aggressive Filtering: Applies aggressive filtering to remove edge cases.
Molecule Filters
Molecule Filters
Specify filters based on RDKit molecular descriptors to keep small-molecule generation within your desired molecular/physicochemical space. Preset filters are available for convenience.
Budget
Budget
Sets the maximum number of molecules for generation and scoring. You can pause/stop the screen at any time.
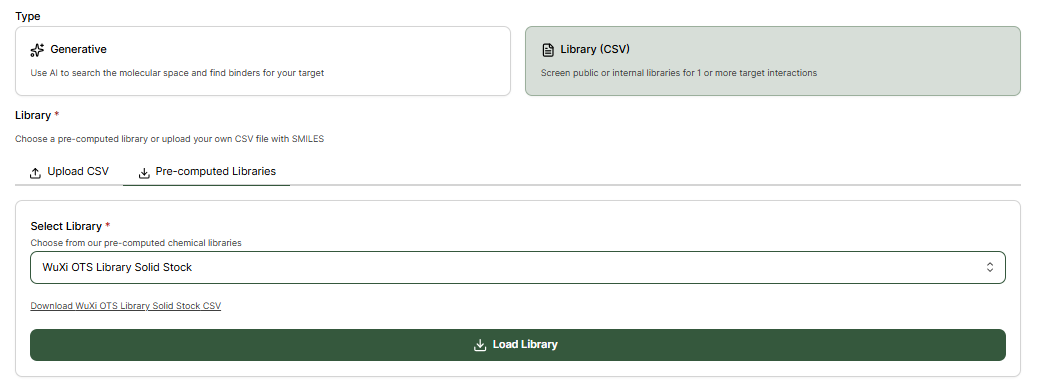
Library Virtual Screen
Custom or Pre-computed Libraries can alternatively be screened by selecting Library (CSV).Upload CSV
The file should contain a SMILES column with the molecules identifier.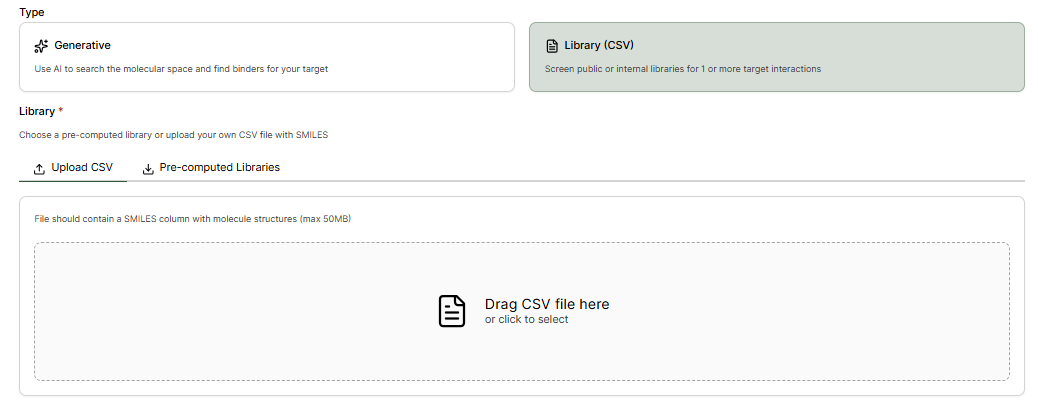
Pre-computed Libraries
Pre-computed vendor compound libraries can be selected without needing to upload CSV files.