Step 1: Create a New Project
- From the home page, click “New Project”
- Enter a project name (e.g., “Designing for external domain of EGFR”)
- Select “Protein” modality
- Click “Create”
Step 2: Add Your First Target
- From the project dashboard, click ”+ Add target” in the Targets section
- Enter target name (e.g., “Target-EGFR”)
- Choose “Define Sequences” tab
- Click ”+ Polymer” then “Protein from Sequence”
- Paste your protein sequence (or import from UniProt)
- Click “Import” then “Continue”
Initialize Target
Complete the 4-step initialization:- Verify Unbound Structure - Review the predicted structure
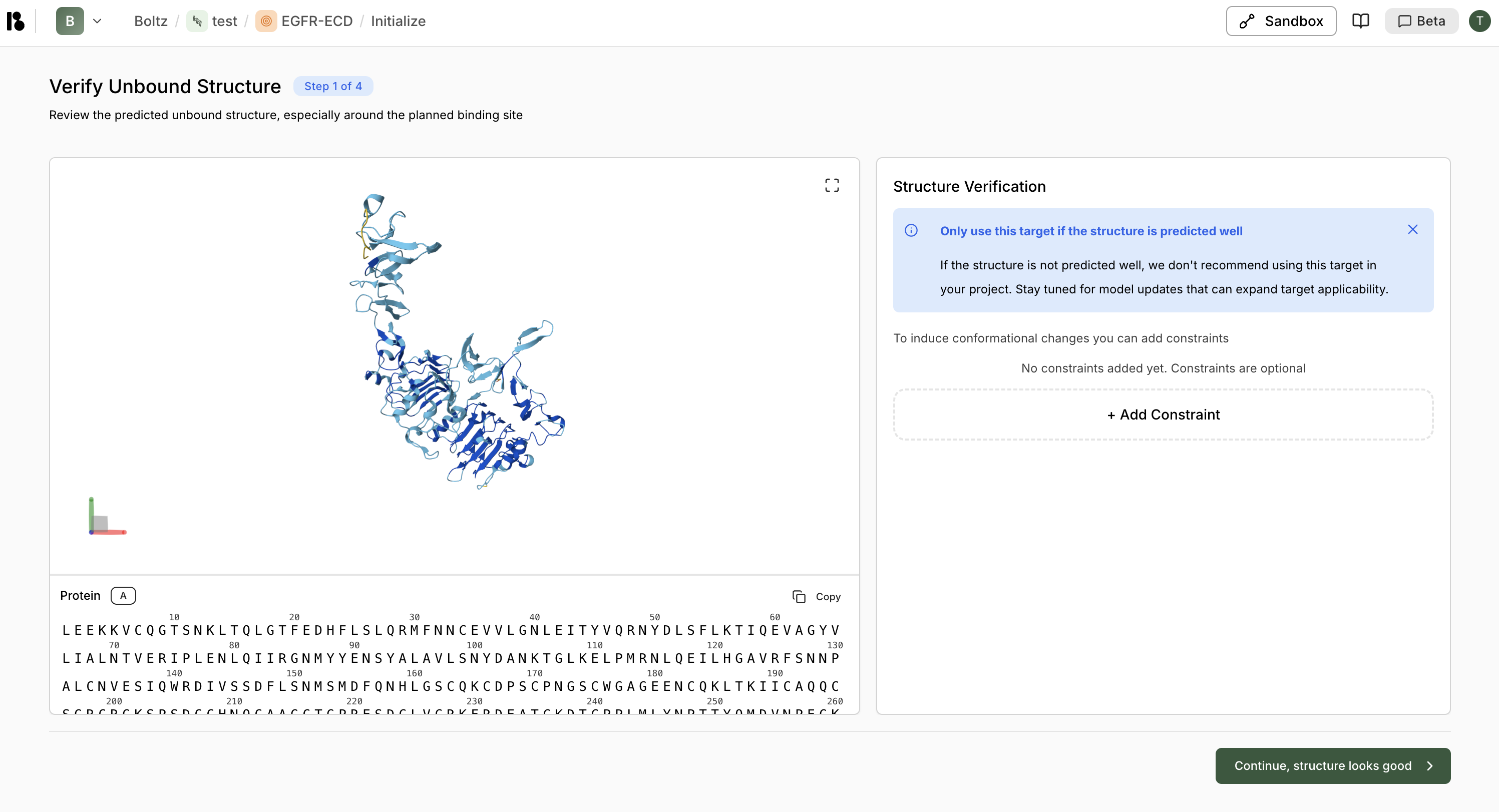
- Select Crop - Choose region of interest (less than 350 residues ideal)
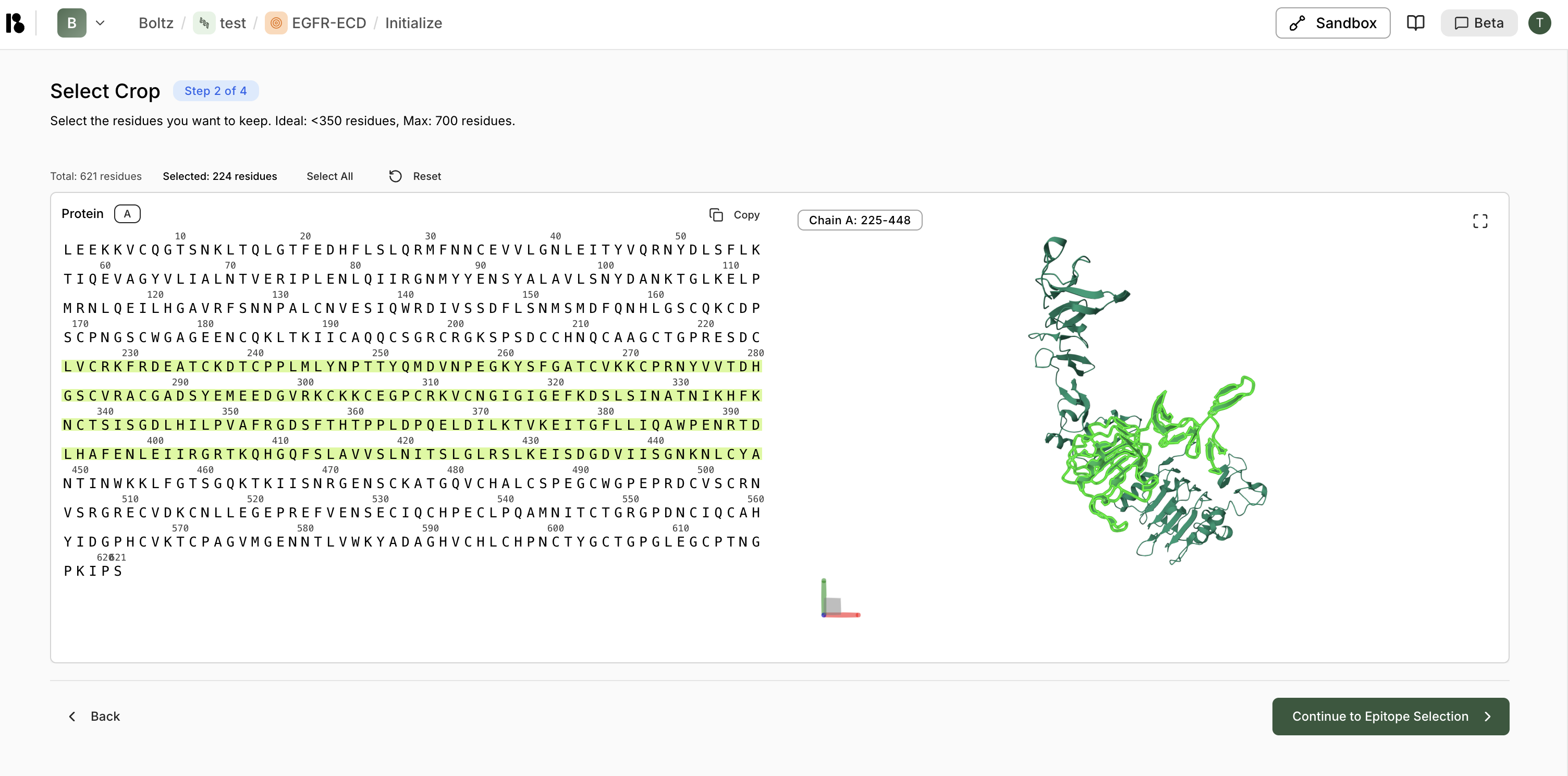
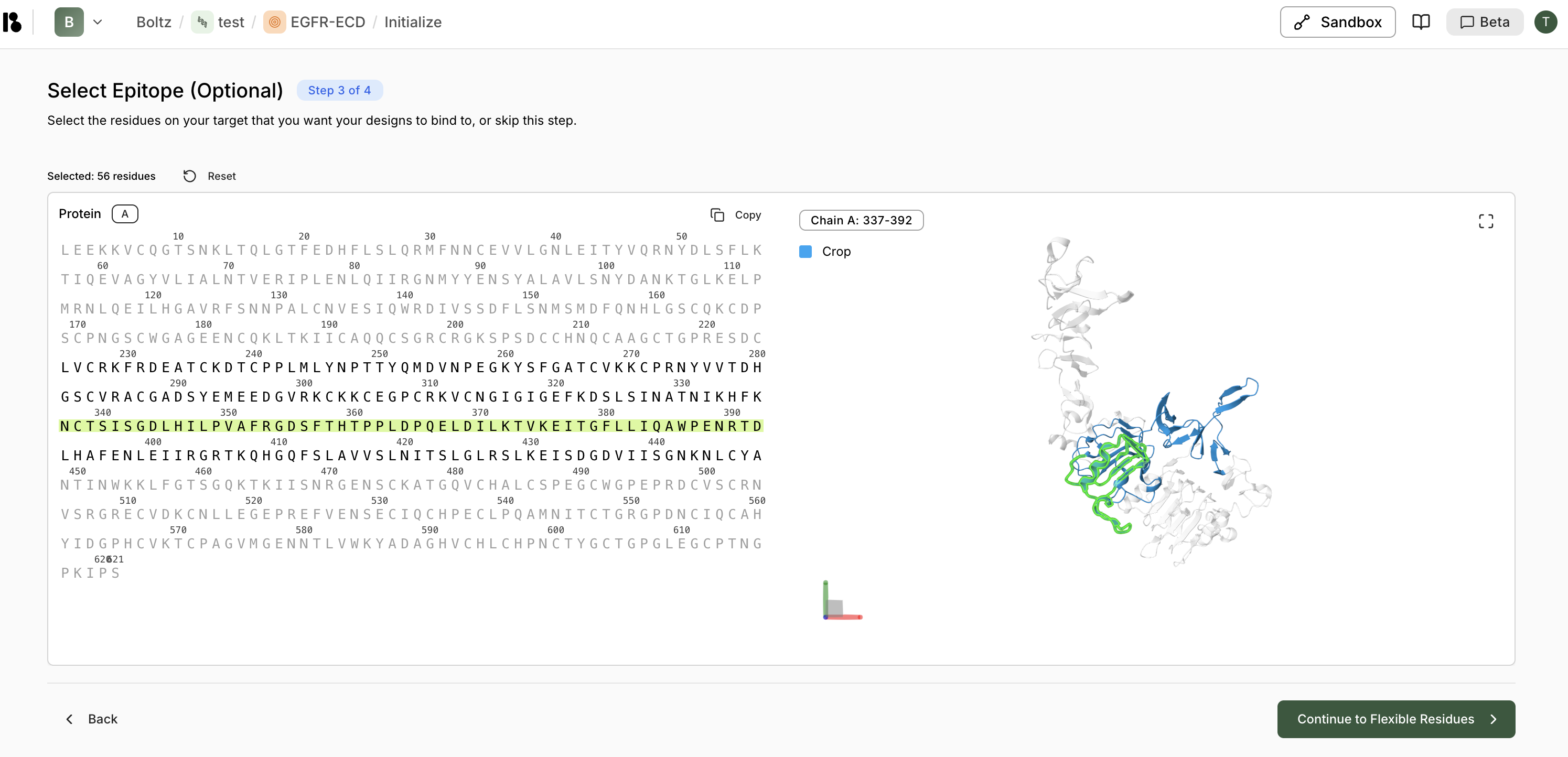
- Select Epitope (Optional) - Mark binding site residues
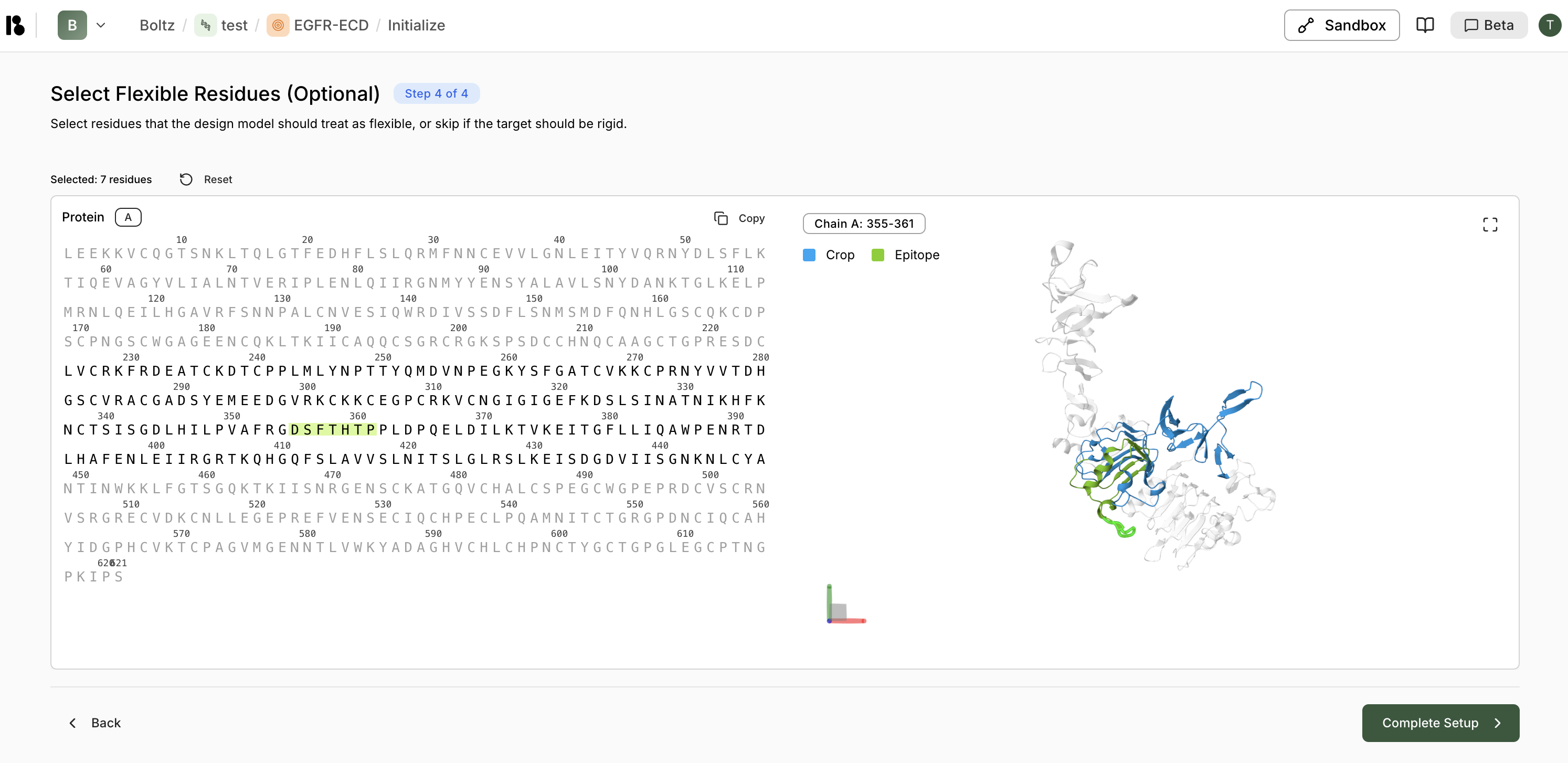
- Select Flexible Residues (Optional) - Mark flexible regions
Step 3: Create a Binder Specification
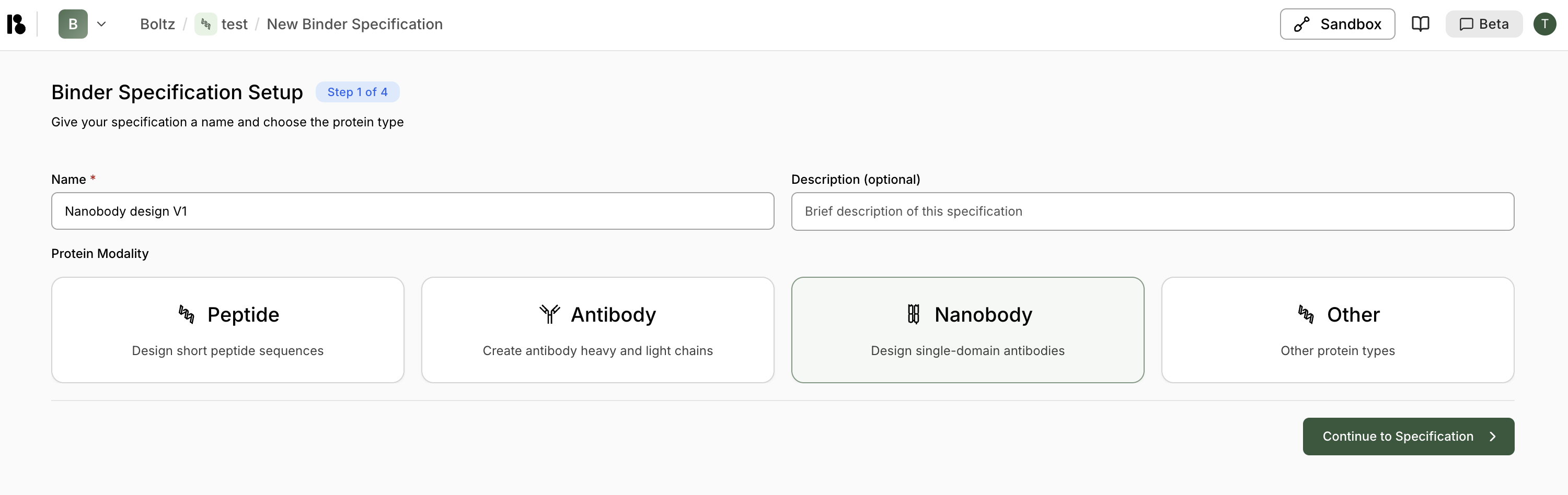
- Click ”+ Add” in the Binder Specifications section
- Enter binder specification name (e.g., “Nanobody design V1”)
- Select “Nanobody” modality
- Click ”> Continue to Specification”
Specify Structure
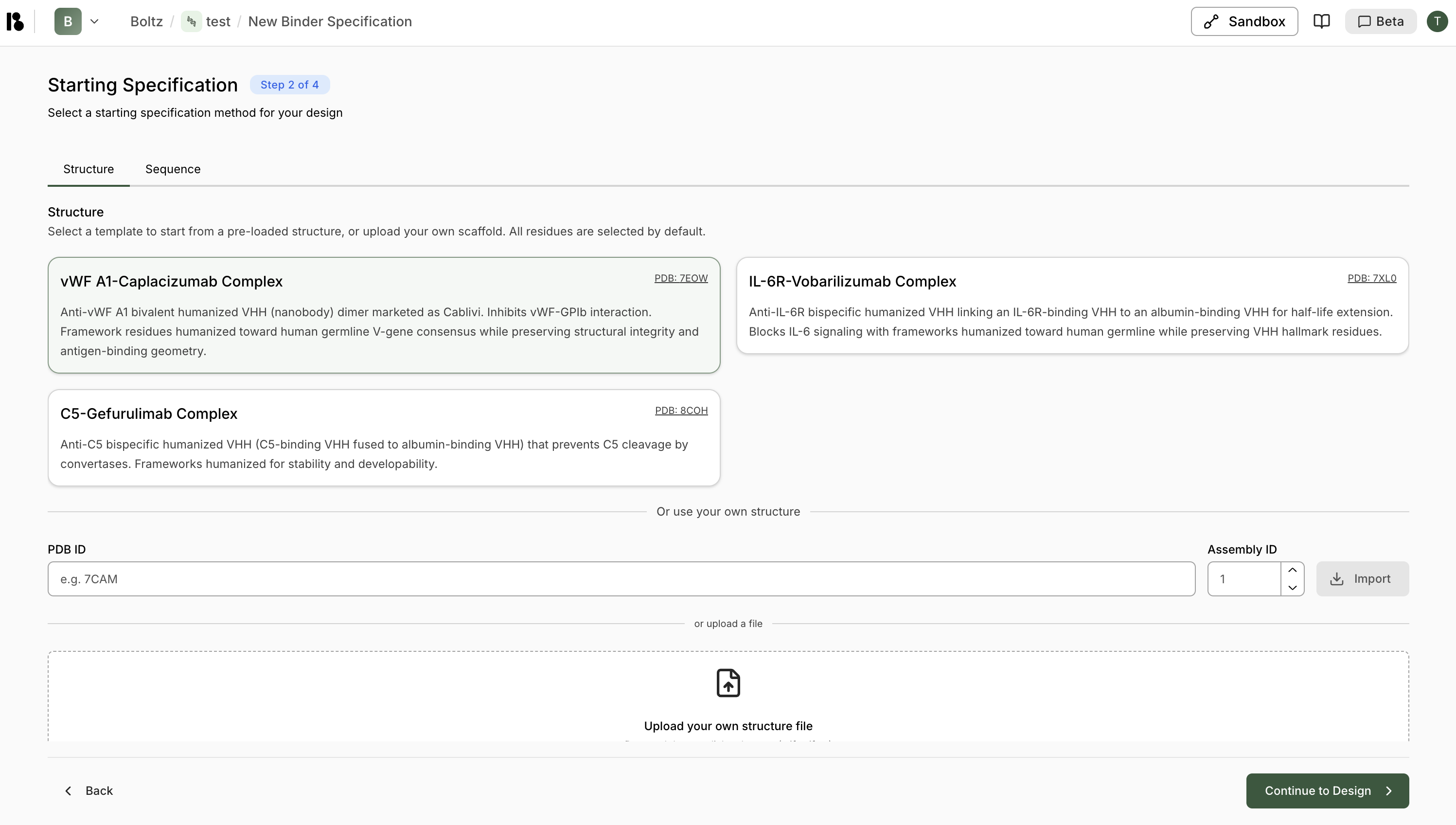
- Pre-loaded template - Select from provided structures e.g., vWF A1-Camplacizumab Complex
- PDB ID - Import via PDB ID (e.g., 7EOW)
- Upload file - Upload your own structure
Select Residues
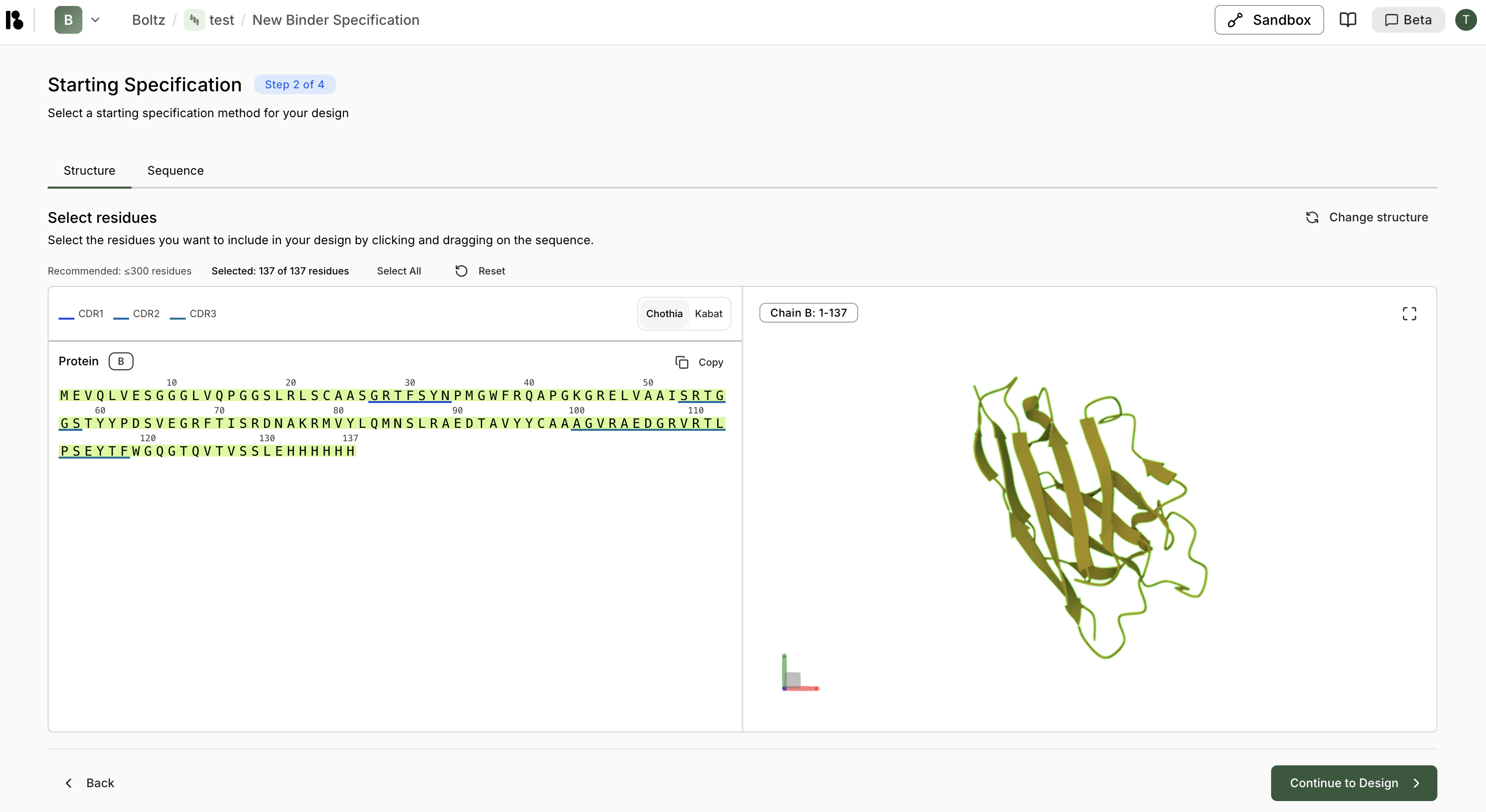
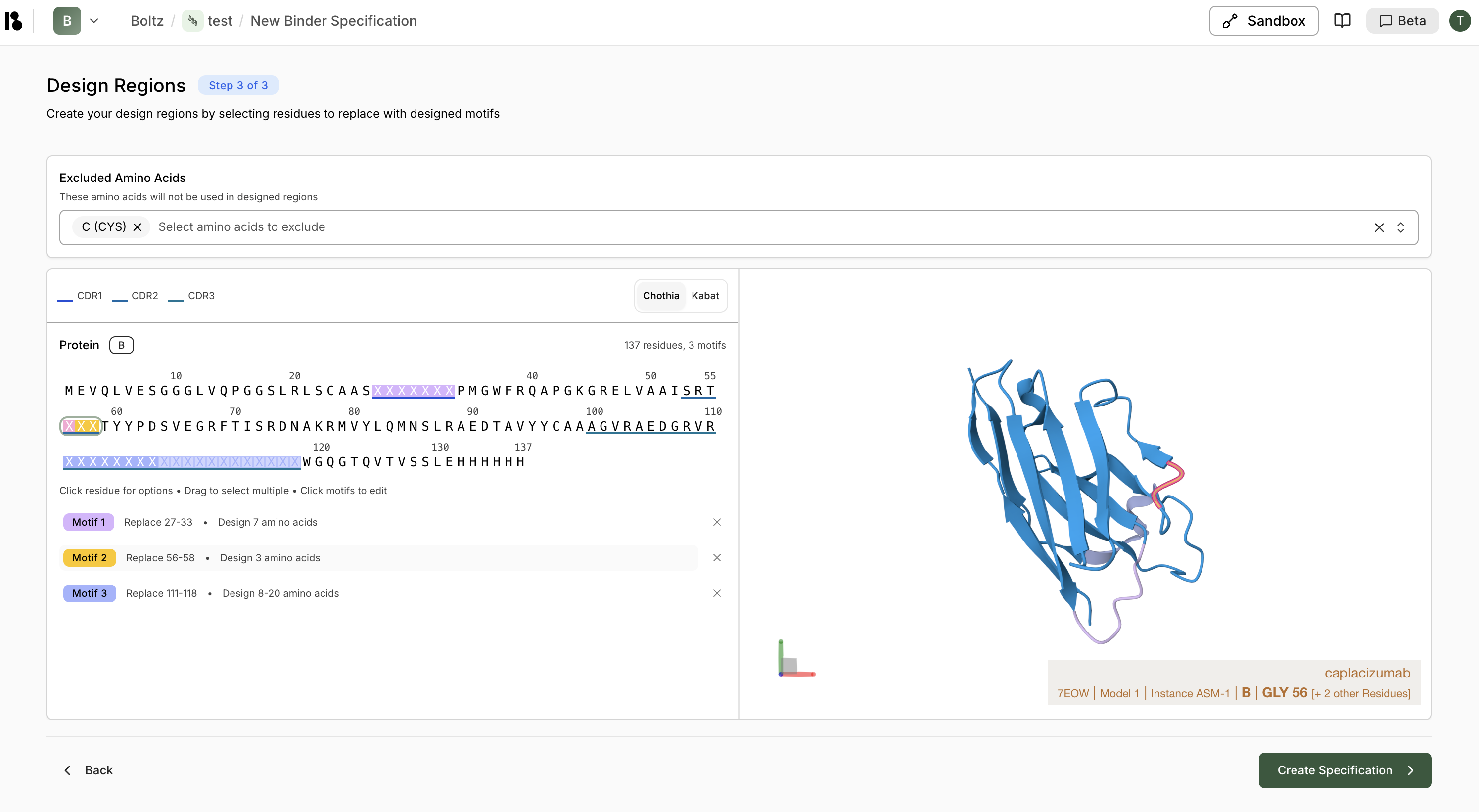
- All residues are selected by default.
- You can also manually select residues by clicking/dragging
- Recommended: ≤300 residues
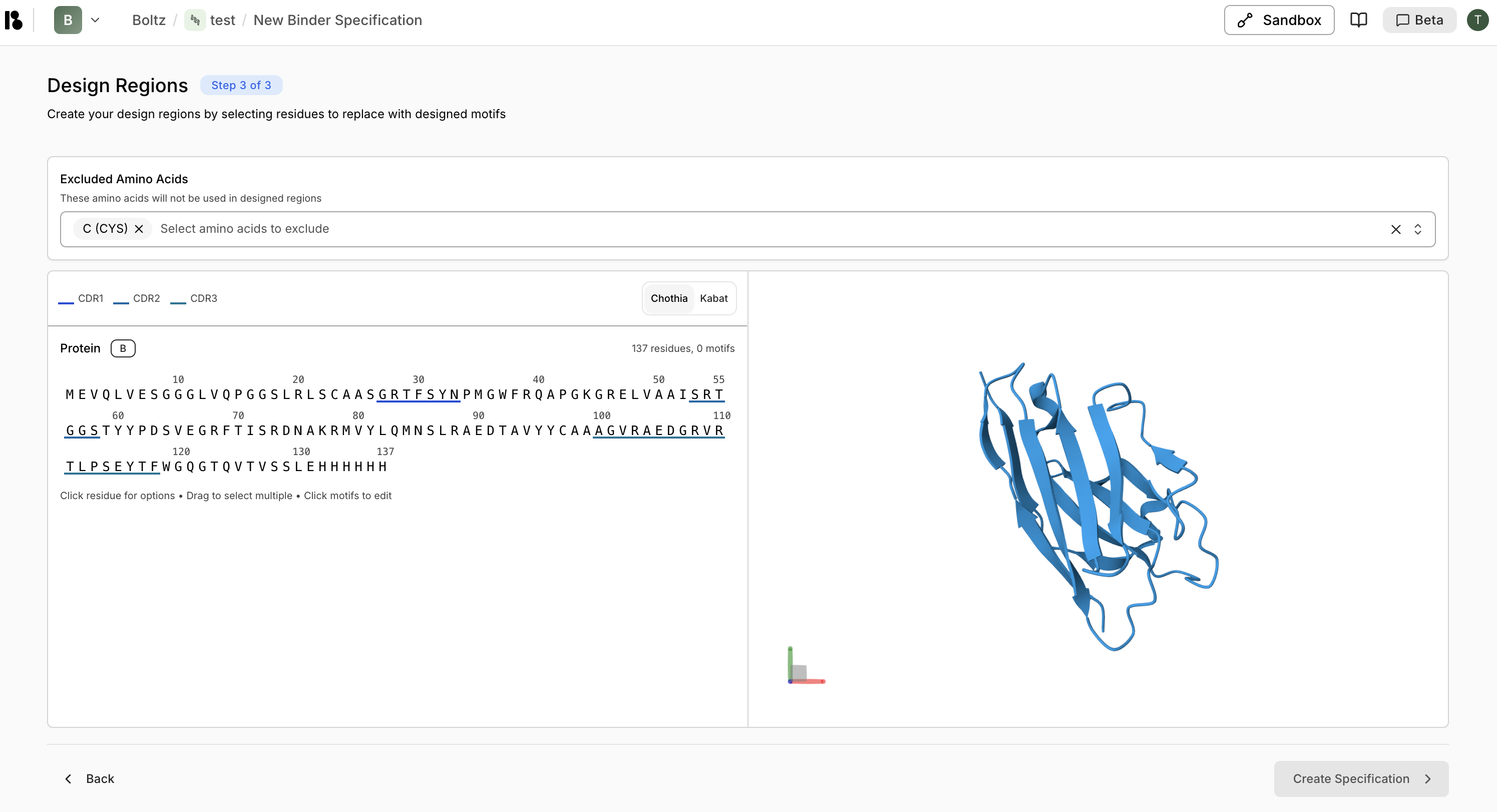
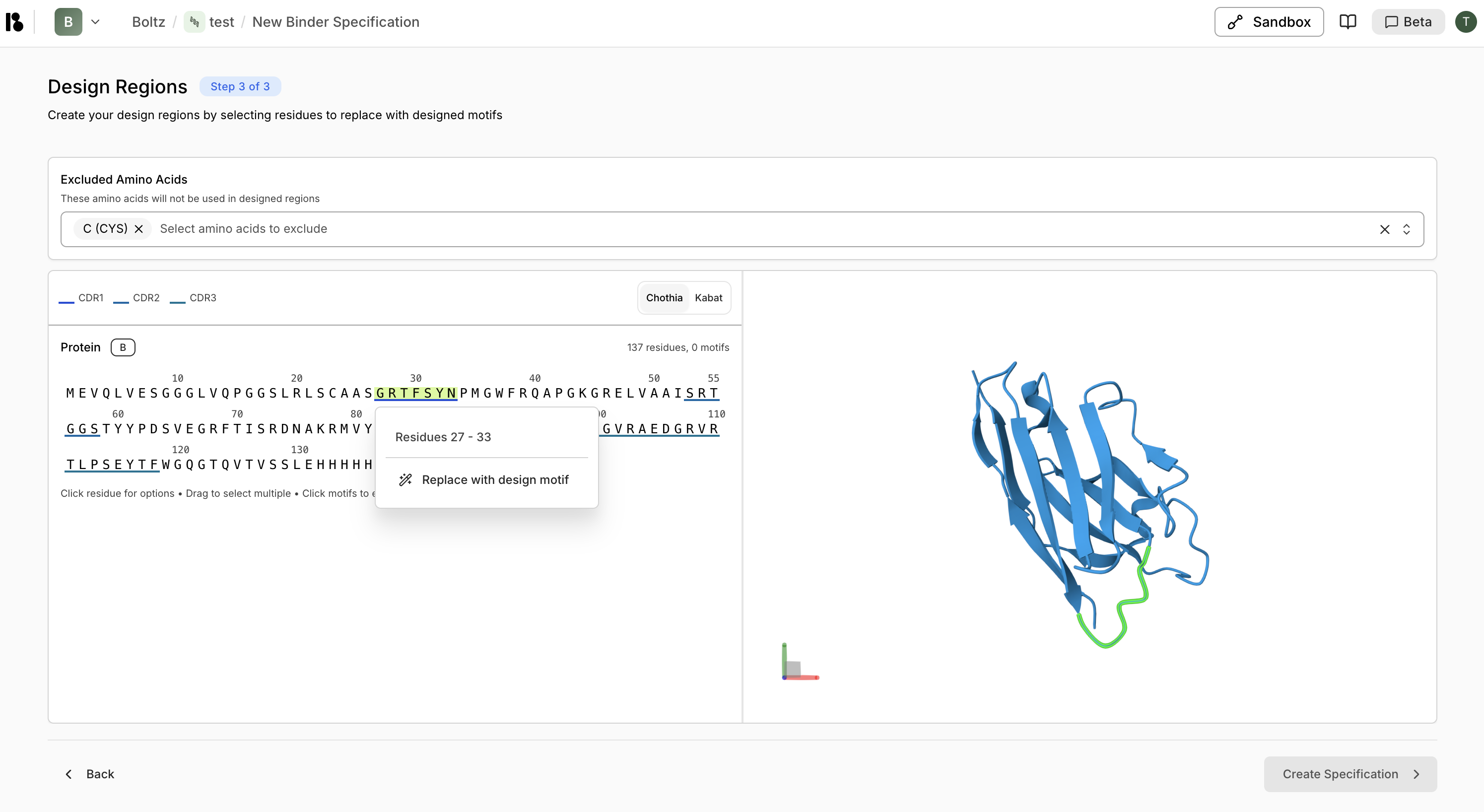
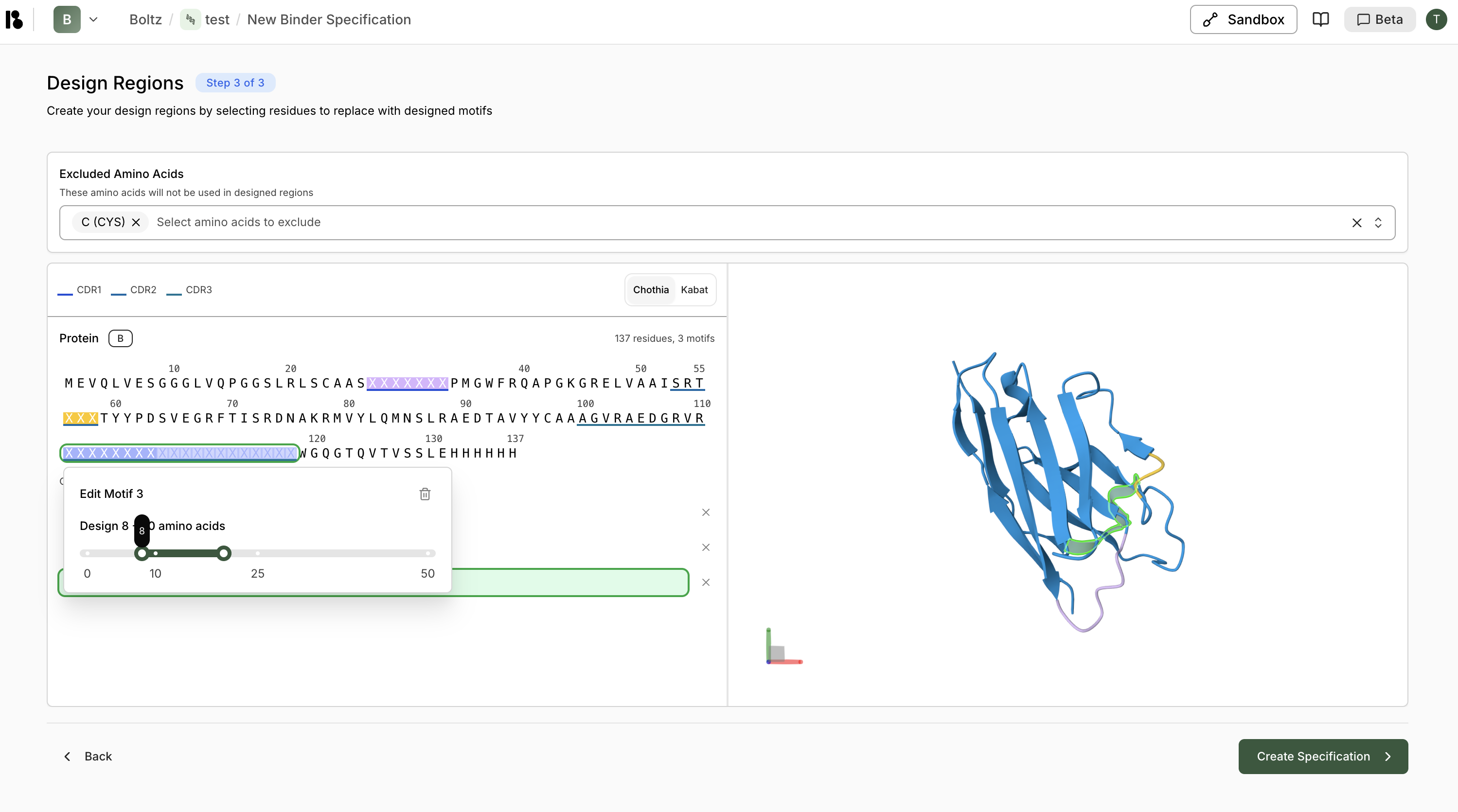
Configure Design
- Exclude amino acids (e.g., Cysteine)
- Create design motifs:
- Select CDR regions
- Right-click then “Replace with design motif”
- For CDR3, specify variable length (e.g., 10-25 residues)
- Click ”> Create Specification”
Step 4: Create an Experiment
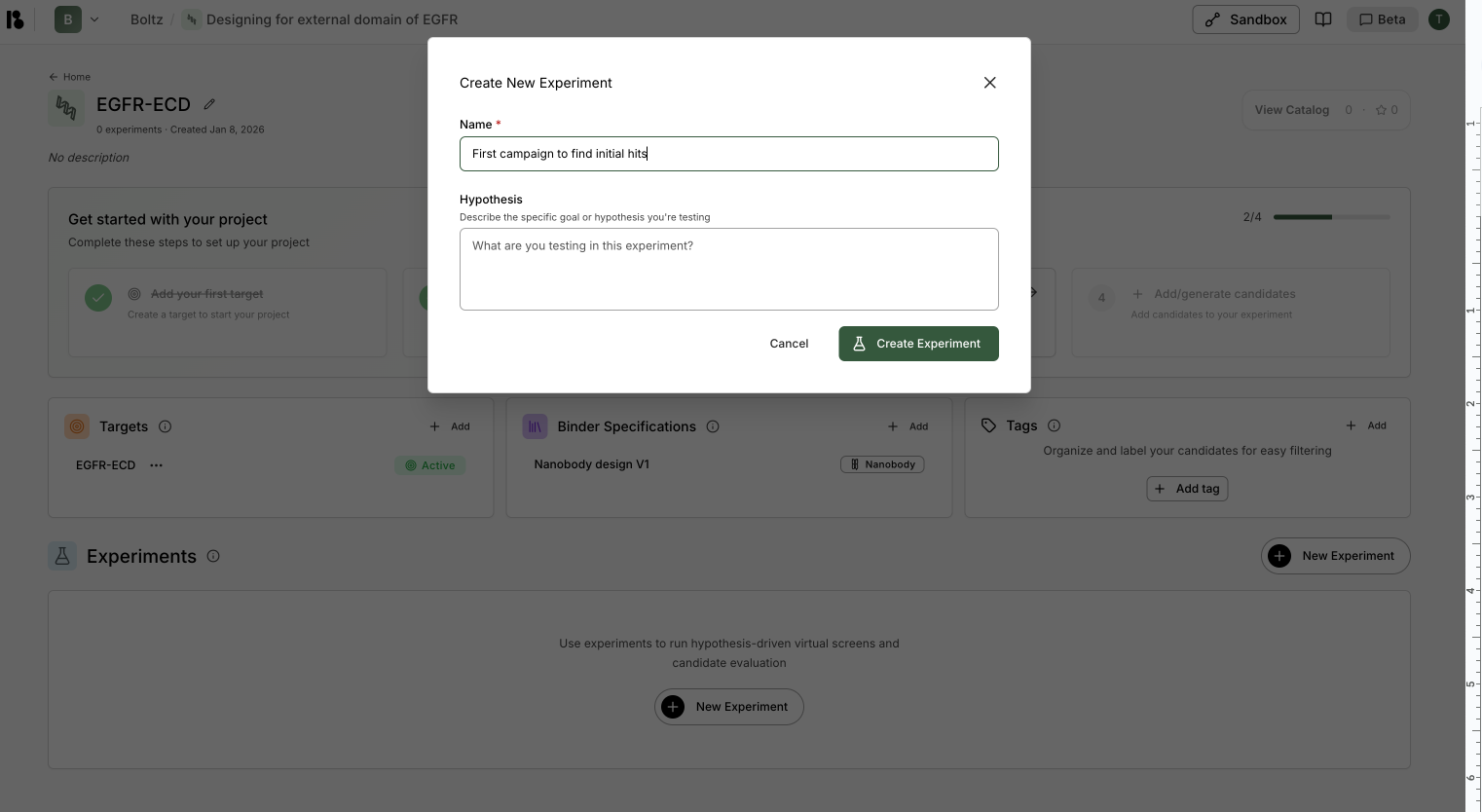
- Click ”+ New Experiment” in the Experiments section
- Enter name (e.g., “First campaign to find initial hits”)
- Enter hypothesis (optional but recommended)
- Click “Create Experiment”
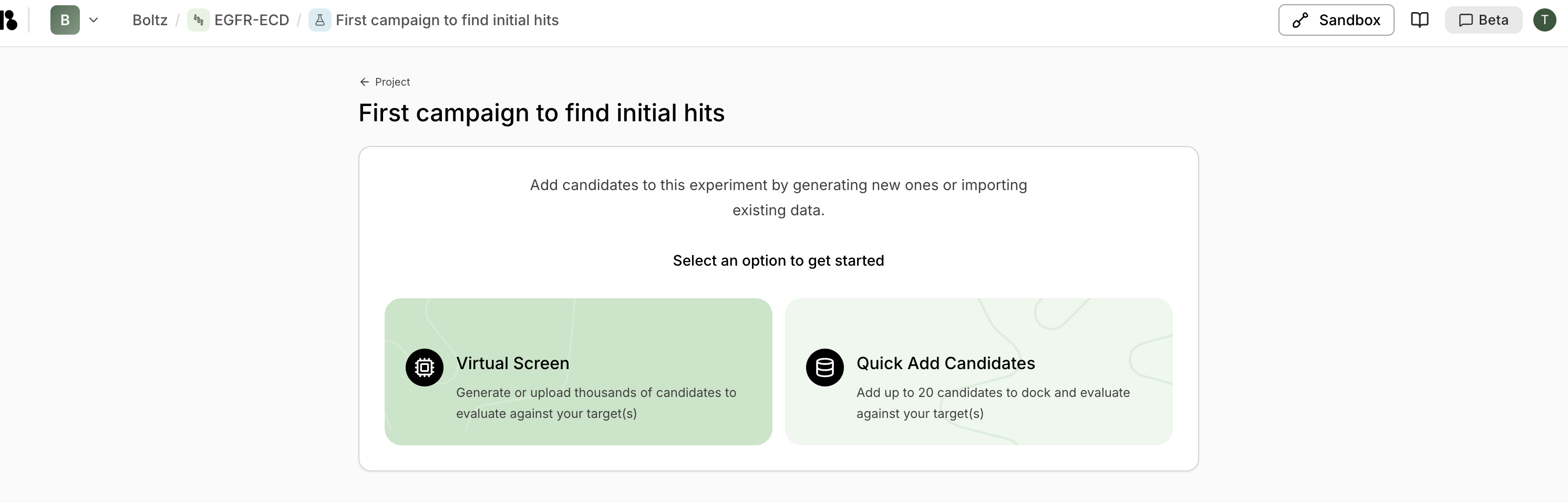
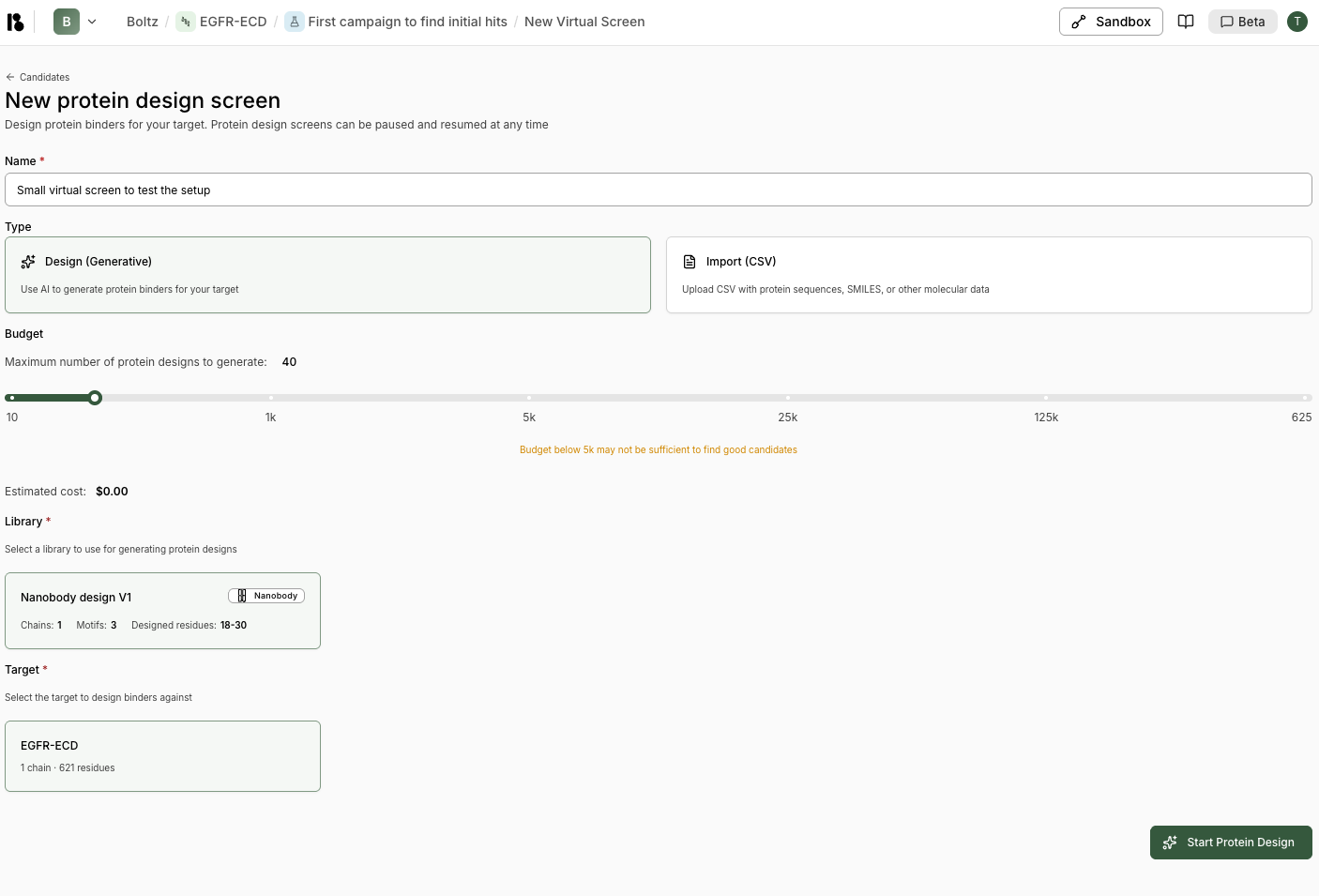
Step 5: Run Virtual Screen
- From the experiment, click “New virtual screen”
- Enter screen name (e.g., “Small virtual screen to test the setup”)
- Select “Design (Generative)” type
- Set budget:
- Start with 30-100 for a pilot
- Scale to 10k-100k for production
- Select your binder specification
- Select your target
- Select your experiment
- Click “Start Protein Design”
Step 6: Monitor Progress
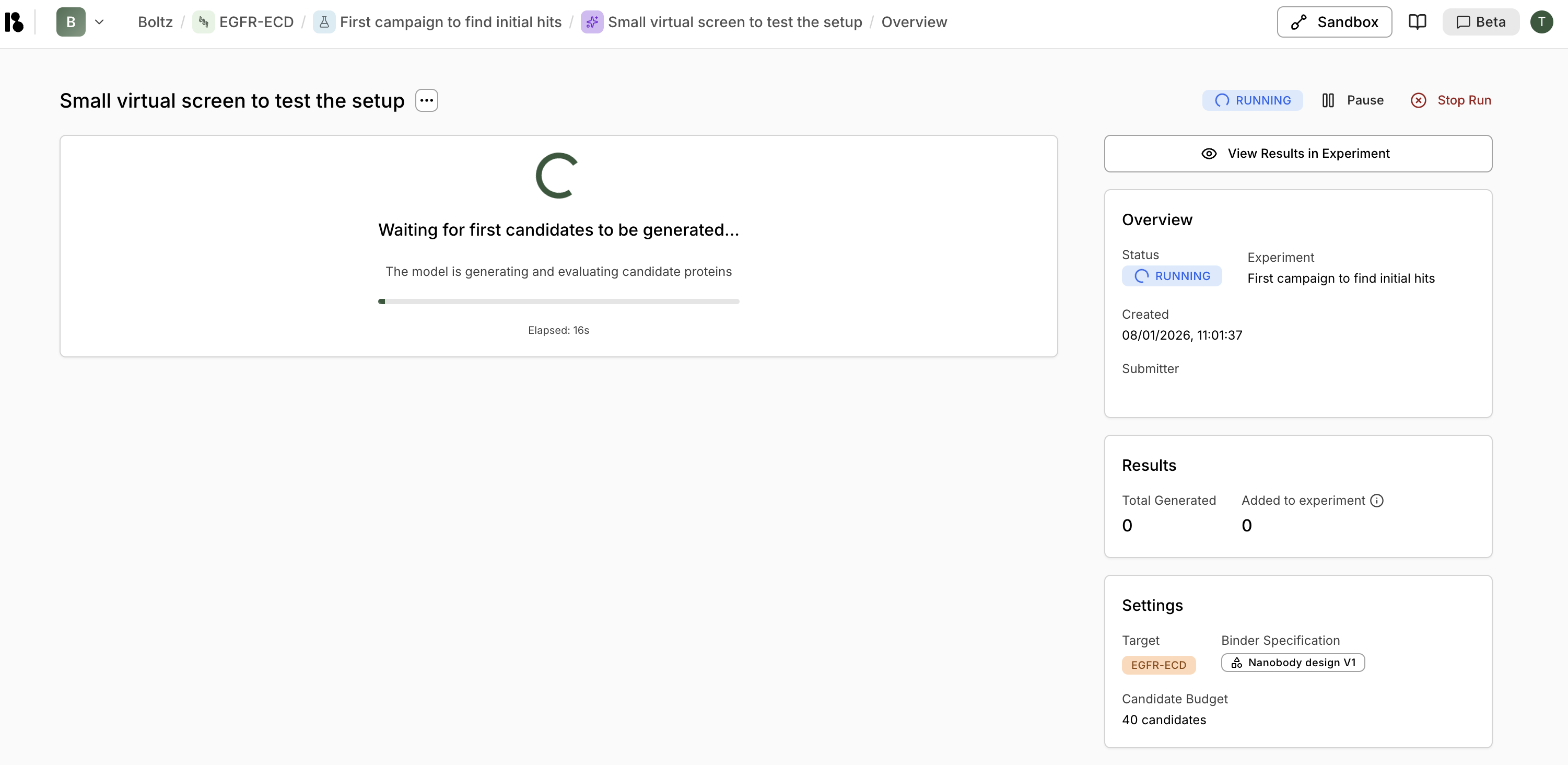
- View progress and elapsed time
- Pause or stop the screen
- Click “View Results in Experiment” to see candidates as they’re generated
Step 7: Evaluate Results
Once completed:- Review metrics:
- Top performers binding confidence graph
- Candidates above threshold counts
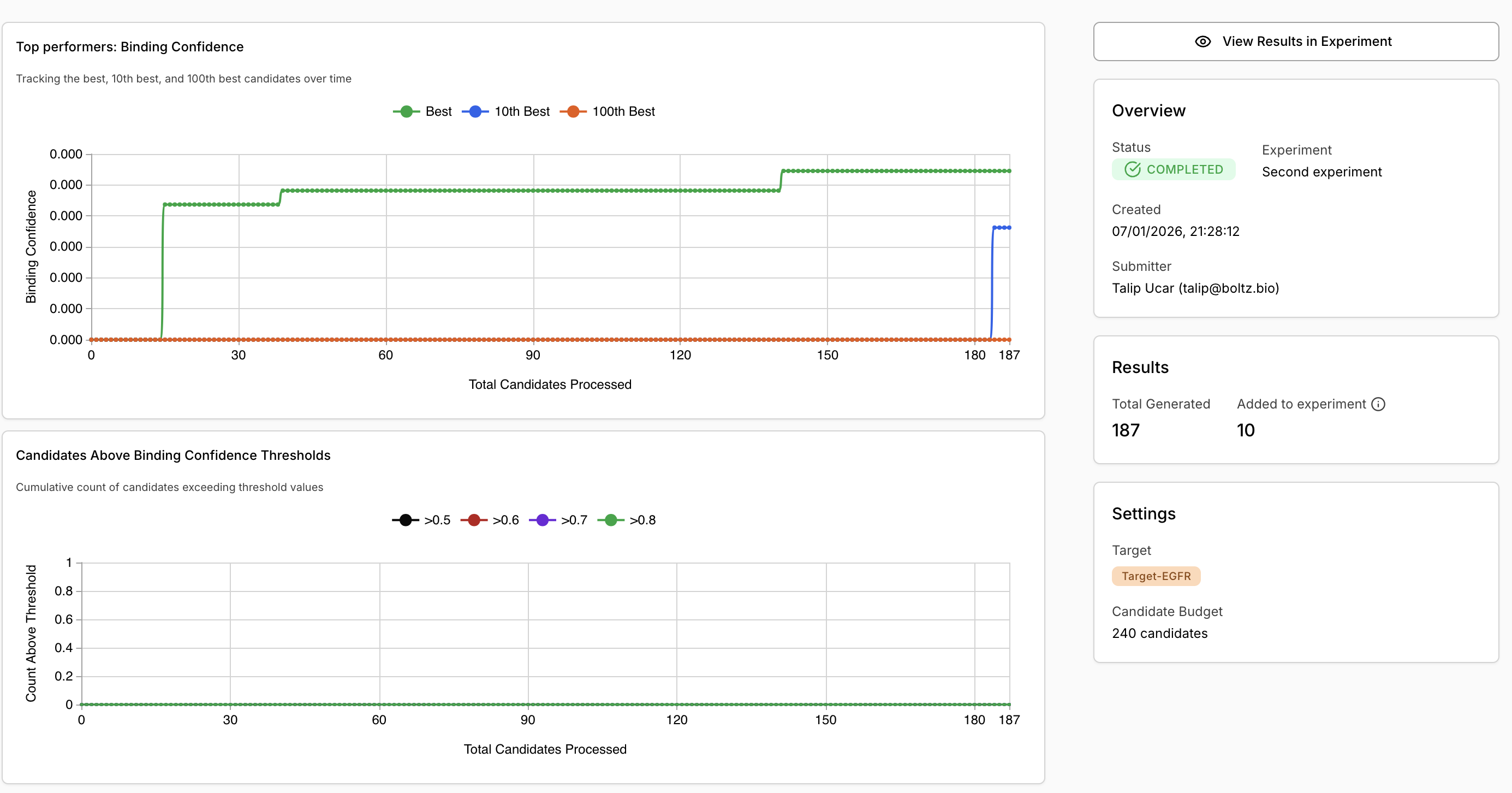
- View candidates:
- Switch to “Table” tab
- Filter and select candidates
- Review 3D structures in the viewer
- Compare properties (ipTM, pLDDT, binding confidence)
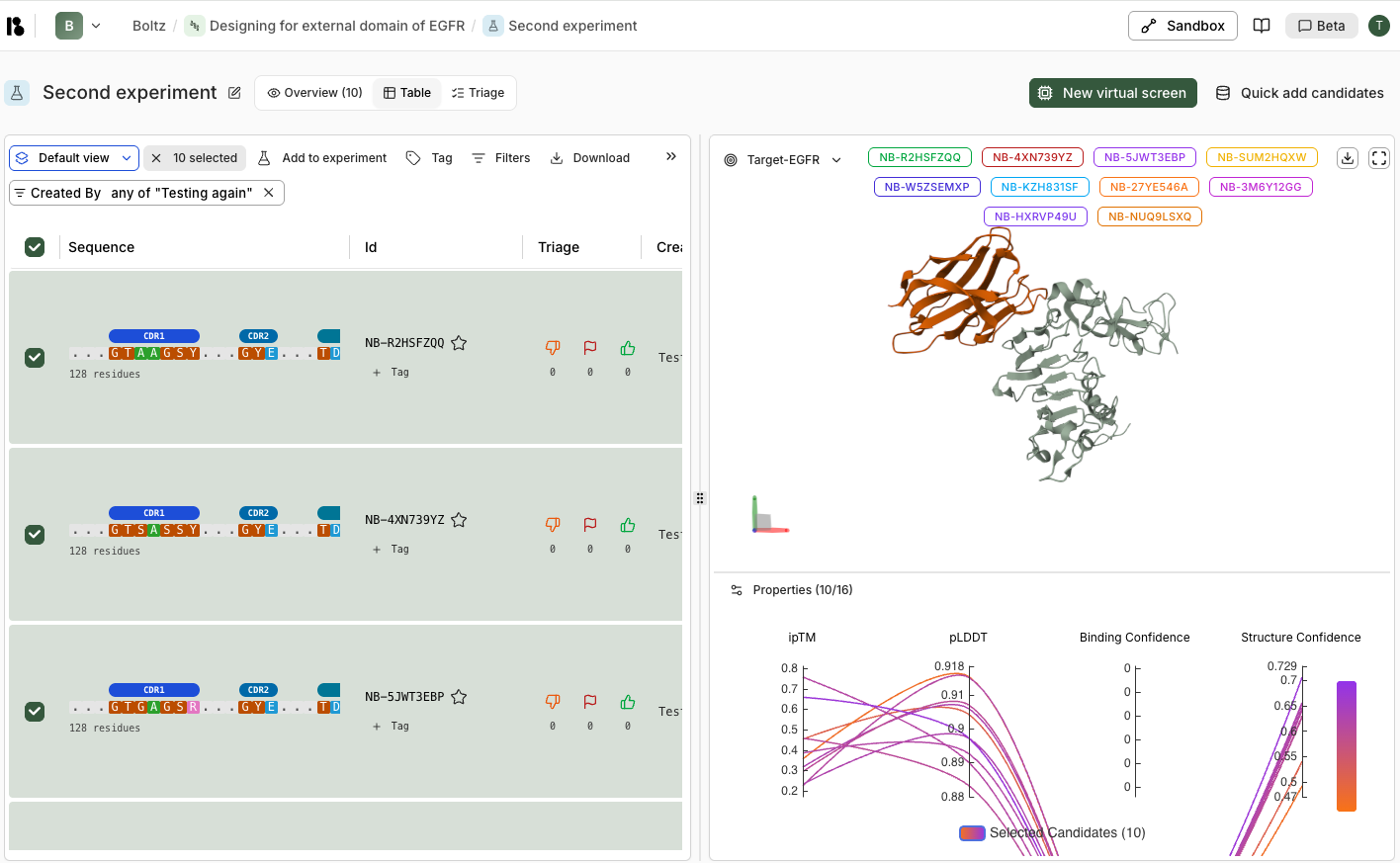
- Triage:
- Use thumbs up/down and flags
- Tag promising candidates
- Export for experimental validation
Best Practices
Start with a small pilot (50-100 designs) to validate setupScale up to 10k-100k designs for production campaignsReview top performers first, then explore diversityUse tags to organize candidates by different criteriaExport promising candidates for experimental validation