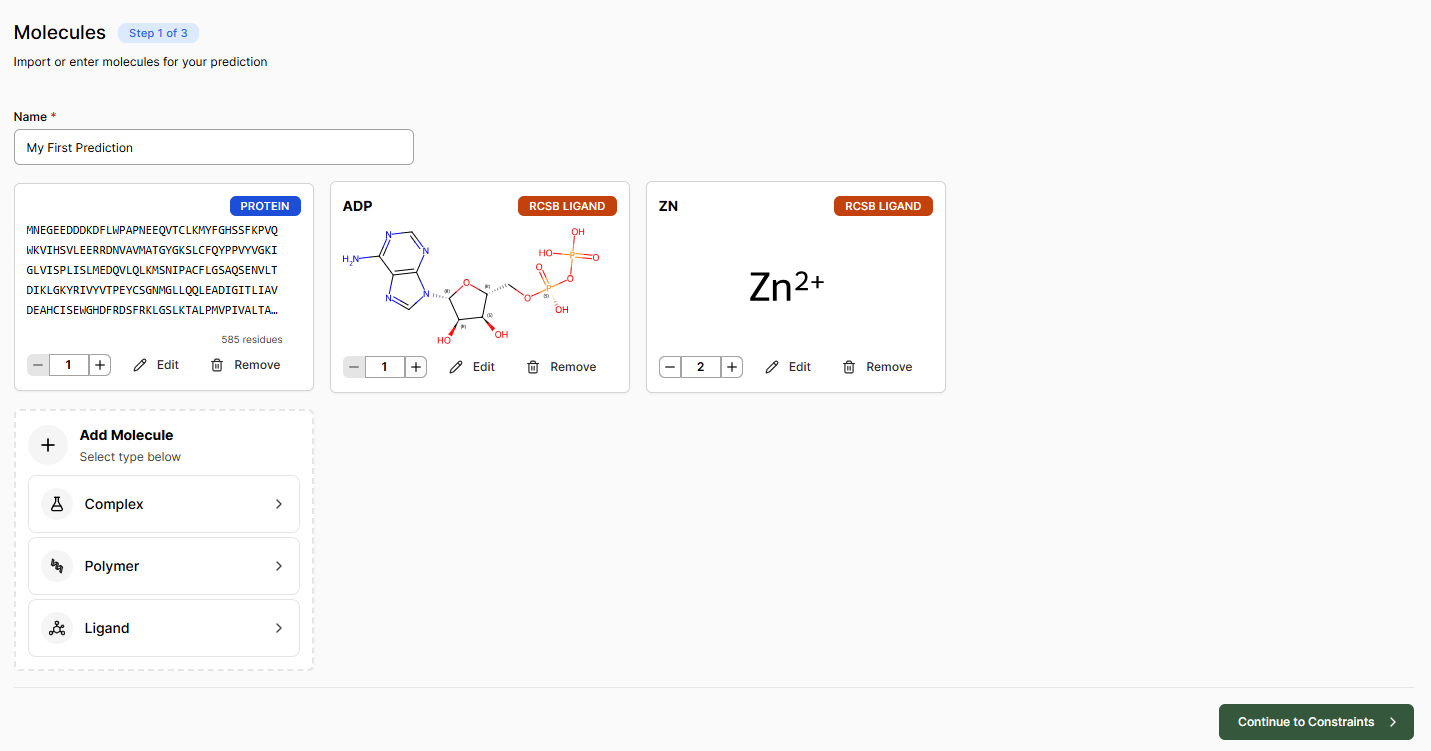
Adding Components
Complex Import
Complex Import
Import a .cif file or provide an RCSB PDB code to load all protein, RNA and DNA sequences, ligands and cofactors present in the file.
Ligand
Ligand
Import small molecules using a SMILES string, the structure drawer, or from an RCSB CCD code.
Polymers
Polymers
Add protein, RNA and DNA sequences individually from UniProt or FASTA strings. Modified amino acids and nucleic acids can be specified when adding a sequence.
Constraints
On the Constraints page, add any bond, contact or pocket-contact information to guide the prediction. Alternatively, skip this step to continue without constraints.| Constraint Type | Description |
|---|---|
| Bond | Specify a covalent bond between atoms in the complex. Important for covalent ligands or cyclic proteins. |
| Contact | Specify a maximum distance (4-20 Å) between a pair of atoms as a hint to the model. |
| Specify residues that a particular binder chain must be within a certain distance from. |
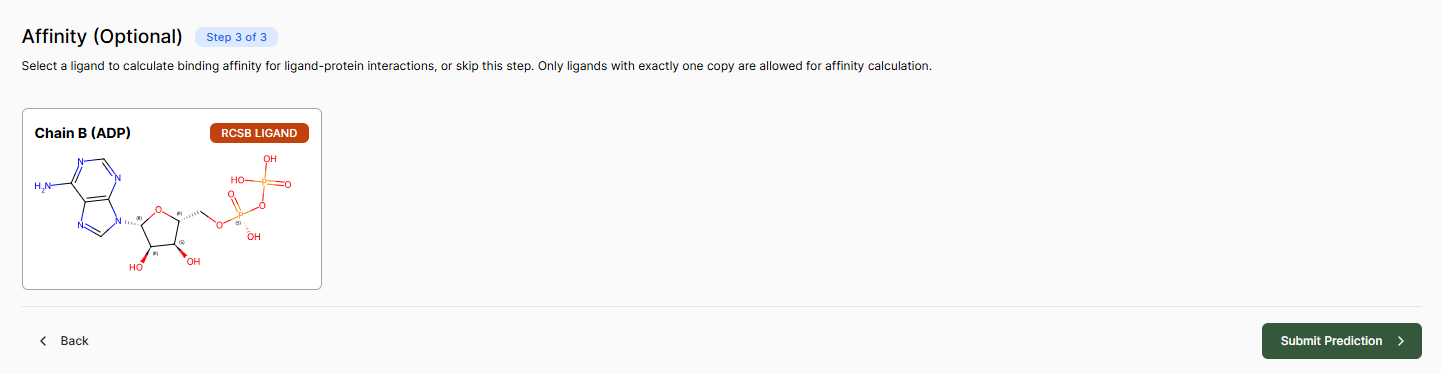
Affinity Prediction
The Affinity page lets you select a ligand to calculate binding affinity for ligand-protein interactions, or skip this step to run a structure-only prediction.