Design Projects are shared workspaces where team members within an organization can collaborate on protein binder design projects. They facilitate hypothesis-driven phases of a research project and provide a space to collate knowledge, designs and predictions in an organized and accessible hierarchy. By default, all work is visible to everyone within your organization.
Project Hierarchy
Design Project
Worked on by a team of scientists, optimizing protein binders against one or more Targets. Contains Experiments as sequential or parallel phases of the design cycle.
Target
The protein structure you want to design binders against. Can be defined by sequence or uploaded structure.
Binder Specifications
Defines the protein binder specifications (e.g., nanobody, antibody, peptide) and design motifs.
Experiment
A hypothesis-driven collection of Virtual Screens used to assess binding poses and affinity scores.
Home
- Organization: Your organization account where you invite members, create API Keys, set up billing, etc.
- Sandbox: A private workspace to test hypotheses and explore protein structures before scaling up.
- CLI: Submit and manage prediction jobs using our command-line interface.
- Projects: All your design projects are listed here.
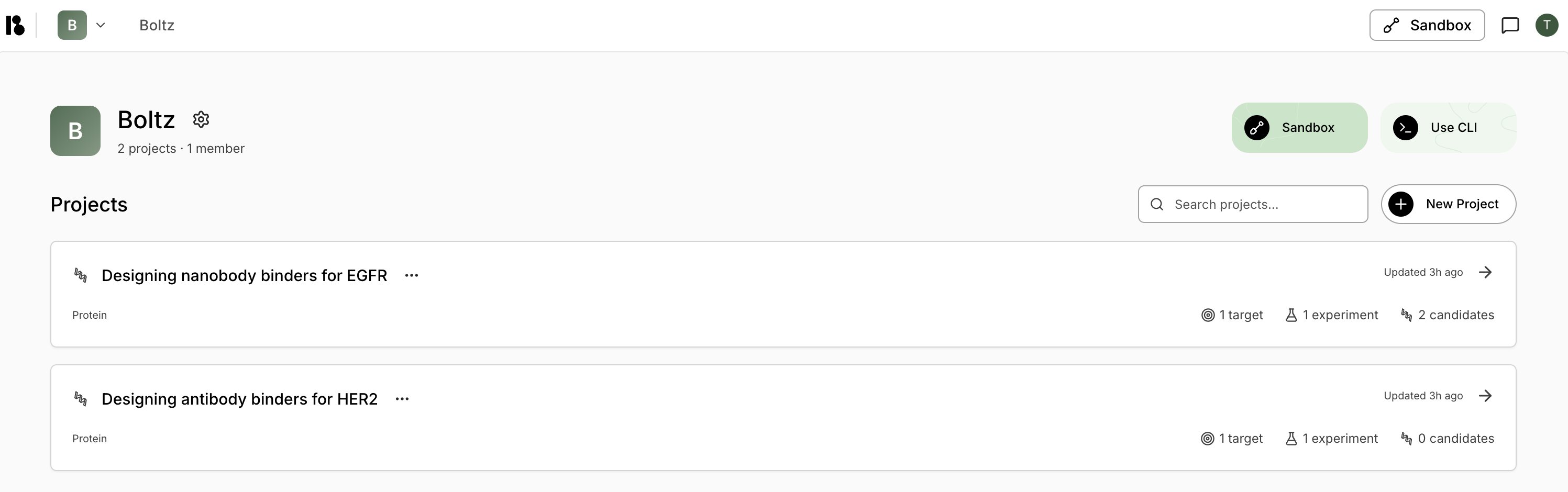
Project Cards
Each project card displays:- Project title and category (Protein/Small Molecule)
- Last updated timestamp
- Metrics: Number of targets, experiments, and candidates
Project Dashboard
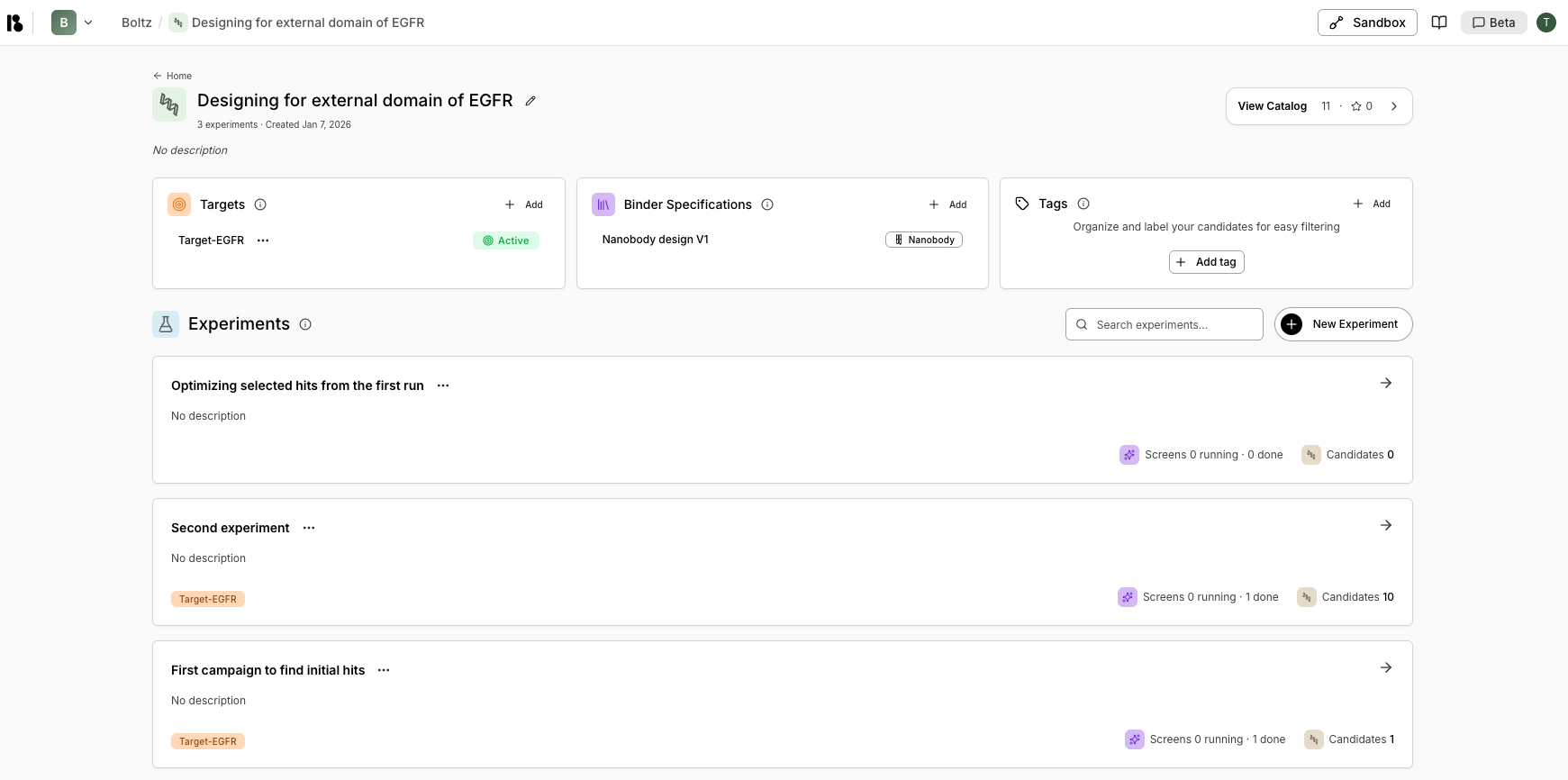
- Targets: List of protein targets added to the project
- Binder Specifications: Defines what proteins you want to design (modality, structure, design regions)
- Tags: Labels used to organize and categorize candidates
- Experiments: All experiments conducted in this project
Getting Started Workflow
New projects include a guided workflow (typically 4 steps):- Add your first target - Create a target to start your project
- Add your binder specification - Define what proteins you want to design
- Create your experiment - Set up an experiment to run virtual screens
- Add/generate candidates - Add candidates to your experiment
Experiment Examples
Good experiment hypotheses are specific and actionable:- Hit Discovery
- Lead Optimization
Name: Designing a de novo protein binder for Pocket A of Target XHypothesis: Identified Pocket A on target X as having potential for blocking target X activity through high-affinity protein binder engagement. Screening 60,000 generated molecules to find the initial hit for a drug discovery campaign.