Introduction
This guide will use the covalent inhibitor AMG 510 bound to KRAS G12C (PDB Code 6OIM) as an example prediction.Modeling of covalent inhibitors is enabled within Boltz Lab, and currently limited to the Sandbox mode.
Submitting a Prediction
Step 1: Create New Prediction
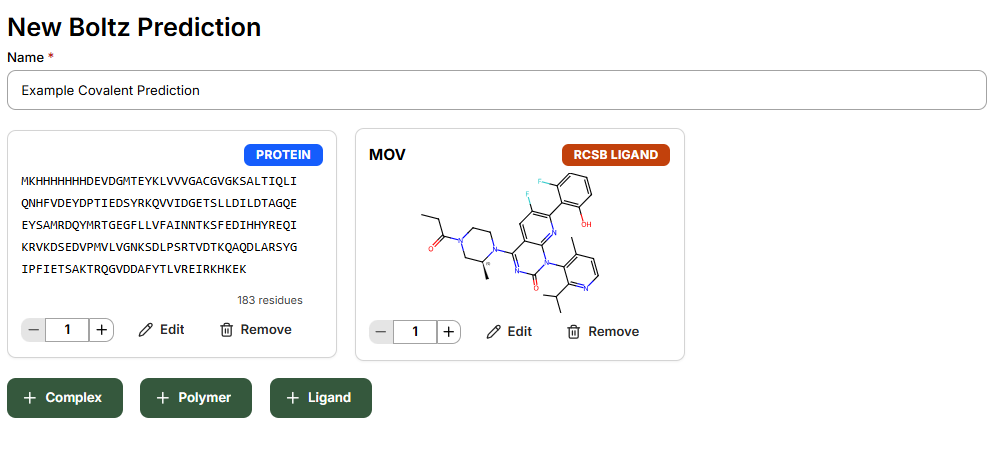
In Sandbox, click New Prediction to get started. After naming your prediction with an identifiable description, add:- Target protein
- Post-modified small molecule
- Any cofactors to be included in the complex
Step 2: Configure Bond Constraint
On the Constraints page, select Bond to guide the prediction. The Bond constraint is used to specify a covalent bond between different atoms in the complex.- Select the Protein residue that you wish the model to covalently modify
- Select the Ligand from the left-hand side
- Ligand: MOV
- Residue number: 26
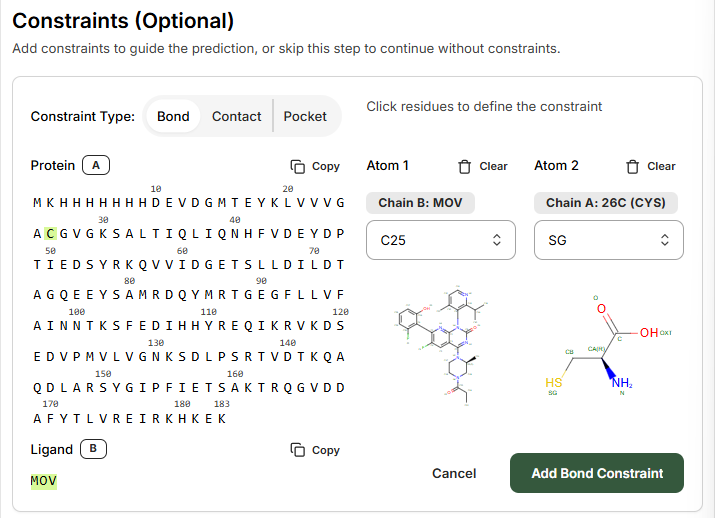
Step 3: Define Atom Connections
Using the right-hand side drop-down menus and atom labeling schematic, select the atoms to be connected. In this example:- Ligand atom: C25
- Protein residue atom: SG
Viewing Results
When inspecting the predicted structure, the covalent bond between ligand and protein is now present.
The prediction results will show the covalent bond formed between the selected atoms, along with standard Boltz prediction metrics including binding likelihood, predicted affinity, and structure confidence scores.
Key Considerations
Warhead Chemistry
Warhead Chemistry
Ensure your input SMILES includes the appropriate warhead structure. For acrylamide-based inhibitors, use the ethyl-substituted amide form rather than the reactive acrylamide.
Residue Selection
Residue Selection
Common covalent modification sites include:
- Cysteine (SG atom)
- Serine (OG atom)
- Lysine (NZ atom)
Current Limitations
Current Limitations
Covalent inhibitor modeling is currently available only in Sandbox mode. Design Projects support for covalent inhibitors is planned for future releases.