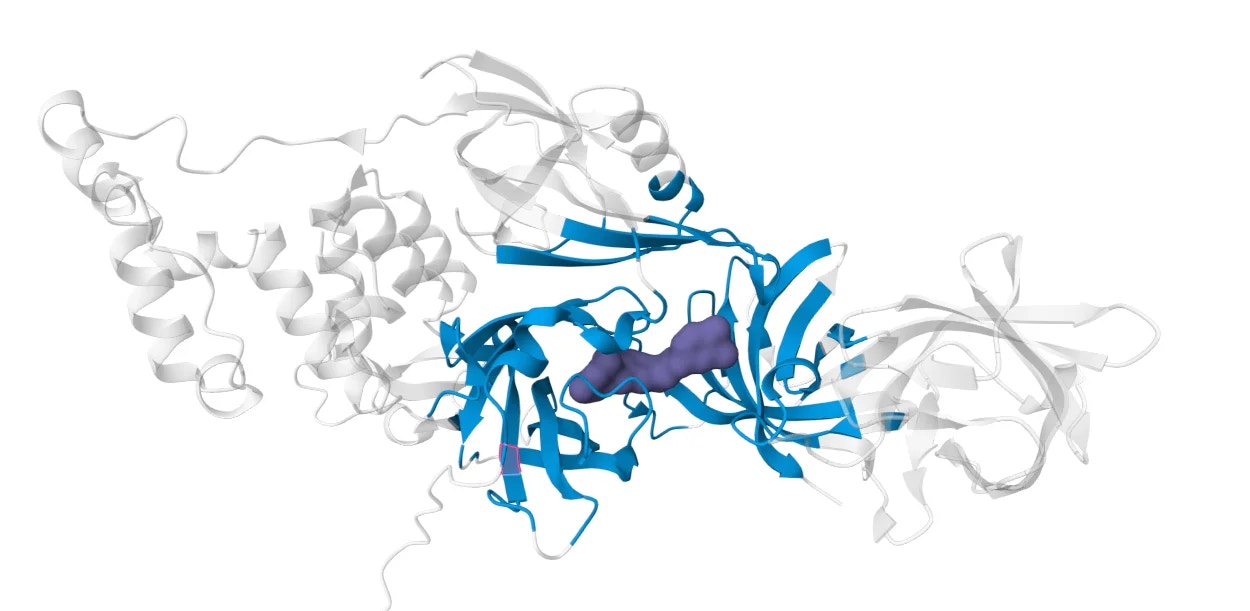
Introduction
Molecular glues function by inducing or stabilizing protein-protein interactions, forming ternary complexes between a target protein, the glue molecule, and a binding partner (such as an E3 ligase or other effector protein). Boltz Lab can facilitate modeling of these ternary complexes within the Sandbox and Design Projects by allowing you to specify multiple protein sequences. This guide uses PDB entry 5HXB as a reference example of a molecular glue ternary complex.Creating a Molecular Glue Target
Step 1: Set Up Design Project
When you create a Target within a Design Project, add two protein sequences instead of one:- Protein 1: Your target protein (e.g., degradation target)
- Protein 2: Binding partner (e.g., E3 ligase, effector protein)
Step 2: Define the Binding Site
There are two approaches to defining where the molecular glue binds:- Pseudo-Pocket (Straightforward)
- Manual Residue Selection
For well-formed interface pockets like PDB 5HXB, the pocket prediction will automatically identify the binding site at the protein-protein interface.This works when:
- The proteins have a clear interface cavity
- The glue binding site is well-defined
- Interface residues are pre-positioned for binding
Guiding Ternary Complex Formation
Using Contact Constraints
If the predicted ternary complex does not match your expected orientation, use the Contact Constraint to guide the model. The Contact constraint defines specific residue-residue interactions between the two proteins, helping to position them correctly relative to each other. When to use Contact constraints:- Proteins are predicted in the wrong relative orientation
- Known key interface contacts are not formed
- The glue binding site is not properly formed between proteins
Contact constraints work by specifying pairs of residues that should be in close proximity, helping the model to place the two proteins into the correct ternary complex geometry.
Key Considerations
Protein Sequence Selection
Protein Sequence Selection
Choose appropriate protein constructs:
- Use domains that are known to interact (avoid including unstructured regions)
- Match experimental constructs when available
- Consider including known co-factors or modifications
Interface Pocket Quality
Interface Pocket Quality
The quality of interface pocket prediction depends on:
- Clarity of the protein-protein interface
- Presence of pre-existing cavity at the interface
- Degree of induced fit required
Glue Molecule Representation
Glue Molecule Representation
Ensure your input molecule represents the actual glue structure:
- Include all relevant functional groups
- Use appropriate protonation states
- Consider stereochemistry if relevant
Validation Strategies
Validation Strategies
Validate your predictions by:
- Comparing to known ternary complex structures (when available)
- Checking that predicted binding poses position the glue at the interface
- Verifying that key interface contacts are maintained
- Confirming predicted binding affinity is reasonable